7ZEW
 
 | | Complex Cyp33-RRM : AAUAAA RNA | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E, RNA (5'-R(*AP*AP*UP*AP*AP*A)-3') | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2023-05-03 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
7ZEV
 
 | | Free form of extended Cyp33-RRM | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2023-05-03 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
7ZEX
 
 | | Complex Cyp33-RRMdelta alpha : UAAUGUCG RNA | | Descriptor: | Isoform 3 of Peptidyl-prolyl cis-trans isomerase E, RNA (5'-R(*UP*AP*AP*UP*GP*UP*CP*G)-3') | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-05-04 | | Last modified: | 2023-05-03 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
7ZEY
 
 | | Complex Cyp33-RRM : MLL1-PHD3 | | Descriptor: | MLL cleavage product N320, Peptidyl-prolyl cis-trans isomerase E, ZINC ION | | Authors: | Blatter, M, Allain, F, Meylan, C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | RNA binding induces an allosteric switch in Cyp33 to repress MLL1-mediated transcription.
Sci Adv, 9, 2023
|
|
6L87
 
 | |
7EA1
 
 | | Crystal Structure of Spindlin1 bound to SPINDOC Docpep2 | | Descriptor: | Peptide from Spindlin interactor and repressor of chromatin-binding protein, Spindlin-1 | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2021-03-05 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for SPINDOC-Spindlin1 engagement and its role in transcriptional inhibition
to be published
|
|
7E9M
 
 | | Crystal Structure of Spindlin1 bound to SPINDOC Docpep3 | | Descriptor: | Peptide from Spindlin interactor and repressor of chromatin-binding protein, Spindlin-1 | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2021-03-04 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for SPINDOC-Spindlin1 engagement and its role in transcriptional inhibition
to be published
|
|
2M1R
 
 | | PHD domain of ING4 N214D mutant | | Descriptor: | Inhibitor of growth protein 4, ZINC ION | | Authors: | Blanco, F.J. | | Deposit date: | 2012-12-05 | | Release date: | 2012-12-26 | | Method: | SOLUTION NMR | | Cite: | Functional impact of cancer-associated mutations in the tumor suppressor protein ING4.
Carcinogenesis, 31, 2010
|
|
2MUM
 
 | | Solution structure of the PHD domain of Yeast YNG2 | | Descriptor: | Chromatin modification-related protein YNG2, ZINC ION | | Authors: | Taeb, S, Kaustov, L, Lemak, A, Farhadi, S, Sheng, Y. | | Deposit date: | 2014-09-12 | | Release date: | 2014-12-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PHD domain of Yeast YNG2
To be Published
|
|
2NS2
 
 | | Crystal Structure of Spindlin1 | | Descriptor: | PHOSPHATE ION, Spindlin-1 | | Authors: | Zhao, Q, Qin, L, Jiang, F, Wu, B, Yue, W, Xu, F, Rong, Z, Yuan, H, Xie, X, Gao, Y, Bai, C, Bartlam, M. | | Deposit date: | 2006-11-02 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human spindlin1. Tandem tudor-like domains for cell cycle regulation
J.Biol.Chem., 282, 2007
|
|
2PUY
 
 | |
2RSD
 
 | | Solution structure of the plant homeodomain (PHD) of the E3 SUMO ligase Siz1 from rice | | Descriptor: | E3 SUMO-protein ligase SIZ1, ZINC ION | | Authors: | Shindo, H, Tsuchiya, W, Suzuki, R, Yamazaki, T. | | Deposit date: | 2012-01-12 | | Release date: | 2012-08-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | PHD finger of the SUMO ligase Siz/PIAS family in rice reveals specific binding for methylated histone H3 at lysine 4 and arginine 2
Febs Lett., 586, 2012
|
|
4LLB
 
 | | Crystal Structure of MOZ double PHD finger histone H3K14ac complex | | Descriptor: | Histone H3.1, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
4LKA
 
 | | Crystal Structure of MOZ double PHD finger histone H3K9ac complex | | Descriptor: | Histone H3.1, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-07 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
4LJN
 
 | | Crystal Structure of MOZ double PHD finger | | Descriptor: | Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-05 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
4LK9
 
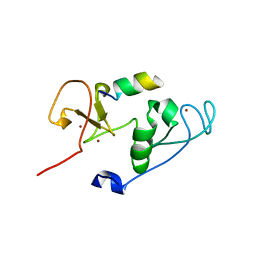 | | Crystal Structure of MOZ double PHD finger histone H3 tail complex | | Descriptor: | Histone H3.1, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-07 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
4FWJ
 
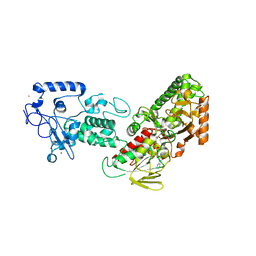 | | Native structure of LSD2/AOF1/KDM1b in spacegroup of I222 at 2.9A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, PHOSPHATE ION, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
4FWE
 
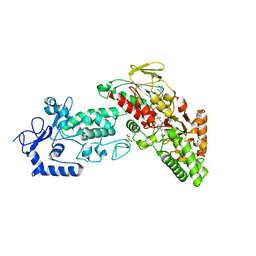 | | Native structure of LSD2 /AOF1/KDM1b in spacegroup of C2221 at 2.13A | | Descriptor: | CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
4FWF
 
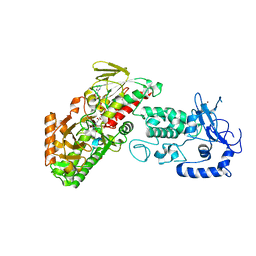 | | Complex structure of LSD2/AOF1/KDM1b with H3K4 mimic | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.1, Lysine-specific histone demethylase 1B, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
4GU0
 
 | | Crystal structure of LSD2 with H3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Yang, H, Dong, Z, Fang, J, Zhu, T, Gong, W, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural insight into substrate recognition by histone demethylase LSD2/KDM1b
Cell Res., 23, 2013
|
|
4GUS
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P3221 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
5VAR
 
 | |
4GUR
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GUT
 
 | | Crystal structure of LSD2-NPAC | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
4GU1
 
 | | Crystal structure of LSD2 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.939 Å) | | Cite: | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
