5MFC
 
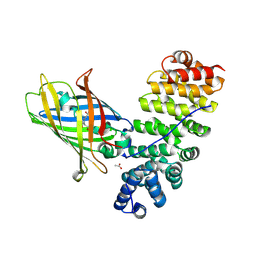 | | Designed armadillo repeat protein YIIIM5AII in complex with (KR)4-GFP | | Descriptor: | (KR)4-Green fluorescent protein,Green fluorescent protein, ACETATE ION, YIIIM5AII | | Authors: | Hansen, S, Kiefer, J, Madhurantakam, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of designed armadillo repeat proteins binding to peptides fused to globular domains.
Protein Sci., 26, 2017
|
|
5B61
 
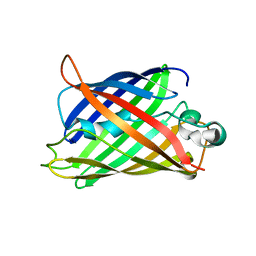 | | Extra-superfolder GFP | | Descriptor: | Green fluorescent protein | | Authors: | Park, H.H, Jang, T.-H, Choi, J.Y. | | Deposit date: | 2016-05-24 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.115 Å) | | Cite: | The mechanism of folding robustness revealed by the crystal structure of extra-superfolder GFP.
FEBS Lett., 591, 2017
|
|
5J3N
 
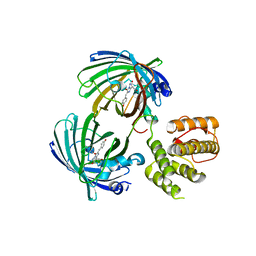 | | C-terminal domain of EcoR124I HsdR subunit fused with the pH-sensitive GFP variant ratiometric pHluorin | | Descriptor: | Green fluorescent protein,HsdR | | Authors: | Grinkevich, P, Iermak, I, Luedtke, N, Mesters, J.R, Ettrich, R, Ludwig, J. | | Deposit date: | 2016-03-31 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of a novel domain of the motor subunit of the Type I restriction enzyme EcoR124 involved in complex assembly and DNA binding.
J. Biol. Chem., 293, 2018
|
|
5LU3
 
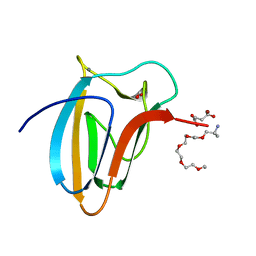 | | The Structure of Spirochaeta thermophila CBM64 | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, 4-oxobutanoic acid, CALCIUM ION, ... | | Authors: | Correia, M.A.S, Romao, M.J, Carvalho, A.L. | | Deposit date: | 2016-09-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stability and Ligand Promiscuity of Type A Carbohydrate-binding Modules Are Illustrated by the Structure of Spirochaeta thermophila StCBM64C.
J. Biol. Chem., 292, 2017
|
|
5HW9
 
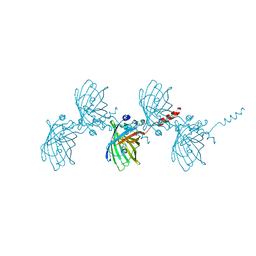 | | Filamentous Assembly of Green Fluorescent Protein Supported by a C-terminal fusion of 18-residues, viewed in space group P21 | | Descriptor: | Green fluorescent protein | | Authors: | Sawaya, M.R, Heller, D.M, McPartland, L, Hochschild, A, Eisenberg, D.S. | | Deposit date: | 2016-01-29 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Green Fluorescent Protein Fusion that Self Assembles as Polar Filaments
to be published
|
|
5WUA
 
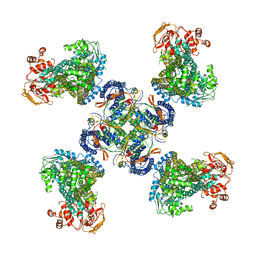 | | Structure of a Pancreatic ATP-sensitive Potassium Channel | | Descriptor: | ATP-sensitive inward rectifier potassium channel 11,superfolder GFP, SUR1 | | Authors: | Li, N, Wu, J.-X, Chen, L, Gao, N. | | Deposit date: | 2016-12-16 | | Release date: | 2017-01-25 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structure of a Pancreatic ATP-Sensitive Potassium Channel
Cell, 168, 2017
|
|
5G5Y
 
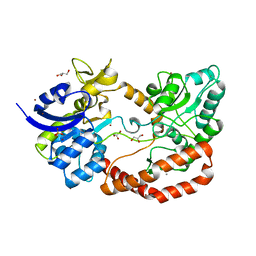 | | S.pneumoniae ABC-transporter substrate binding protein FusA apo structure | | Descriptor: | ABC TRANSPORTER, SUBSTRATE-BINDING PROTEIN, CALCIUM ION, ... | | Authors: | Culurgioni, S, Harris, G, Singh, A.K, King, S.J, Walsh, M.A. | | Deposit date: | 2016-06-10 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis for Regulation and Specificity of Fructooligosaccharide Import in Streptococcus pneumoniae.
Structure, 25, 2017
|
|
5FVG
 
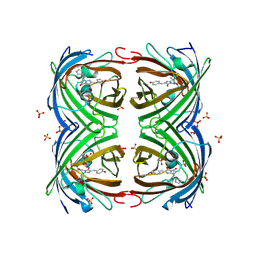 | | Structure of IrisFP at 100 K. | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett, 7, 2016
|
|
5HGE
 
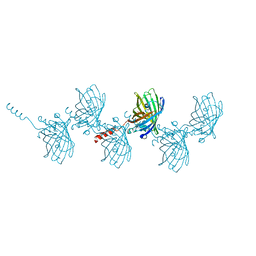 | | Filamentous Assembly of Green Fluorescent Protein Supported by a C-terminal fusion of 18-residues, viewed in space group P212121 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Green fluorescent protein | | Authors: | Sawaya, M.R, Heller, D.M, McPartland, L, Hochschild, A, Eisenberg, D.S. | | Deposit date: | 2016-01-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Green Fluorescent Protein Fusion that Self Assembles as Polar Filaments
to be published
|
|
5HBD
 
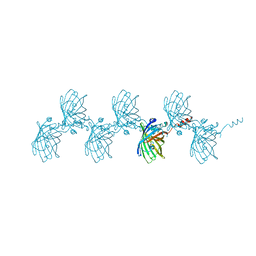 | | Filamentous Assembly of Green Fluorescent Protein Supported by a C-terminal fusion of 18-residues, viewed in space group C2 | | Descriptor: | Green fluorescent protein | | Authors: | Sawaya, M.R, Hochschild, A, Heller, D.M, McPartland, L, Eisenberg, D.S. | | Deposit date: | 2015-12-31 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Green Fluorescent Protein Fusion that Self Assembles as Polar Filaments
to be published
|
|
5LTP
 
 | |
5LTQ
 
 | |
5LTR
 
 | |
5T3I
 
 | |
5K4Z
 
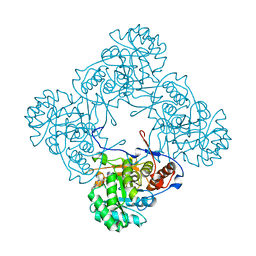 | | M. thermoresistible IMPDH in complex with IMP and Compound 6 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ~{N}-(4-fluorophenyl)-4-(2~{H}-indazol-6-ylsulfamoyl)-3,5-dimethyl-1~{H}-pyrrole-2-carboxamide | | Authors: | Pacitto, A, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-05-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Essential but Not Vulnerable: Indazole Sulfonamides Targeting Inosine Monophosphate Dehydrogenase as Potential Leads against Mycobacterium tuberculosis.
ACS Infect Dis, 3, 2017
|
|
5K4X
 
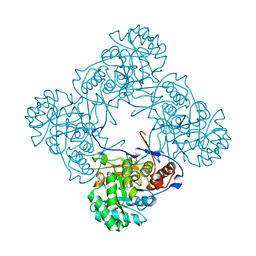 | | M. thermoresistible IMPDH in complex with IMP and Compound 1 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ~{N}-(2~{H}-indazol-6-yl)-3,5-dimethyl-1~{H}-pyrazole-4-sulfonamide | | Authors: | Pacitto, A, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-05-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Essential but Not Vulnerable: Indazole Sulfonamides Targeting Inosine Monophosphate Dehydrogenase as Potential Leads against Mycobacterium tuberculosis.
ACS Infect Dis, 3, 2017
|
|
3JBM
 
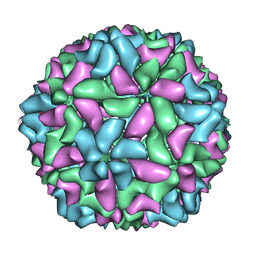 | | Electron cryo-microscopy of a virus-like particle of orange-spotted grouper nervous necrosis virus | | Descriptor: | virus-like particle of orange-spotted grouper nervous necrosis virus | | Authors: | Xie, J, Li, K, Gao, Y, Huang, R, Lai, Y, Shi, Y, Yang, S, Zhu, G, Zhang, Q, He, J. | | Deposit date: | 2015-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural analysis and insertion study reveal the ideal sites for surface displaying foreign peptides on a betanodavirus-like particle
Vet. Res., 47, 2016
|
|
5K9D
 
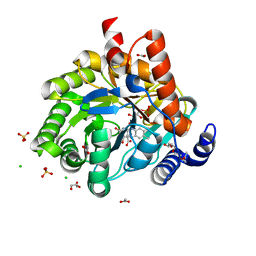 | | Crystal structure of human dihydroorotate dehydrogenase at 1.7 A resolution | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Lewis, T.A, Sykes, D.B, Law, J.M, Munoz, B, Scadden, D.T, Rustiguel, J.K, Nonato, M.C, Schreiber, S.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of ML390: A Human DHODH Inhibitor That Induces Differentiation in Acute Myeloid Leukemia.
ACS Med Chem Lett, 7, 2016
|
|
5K9C
 
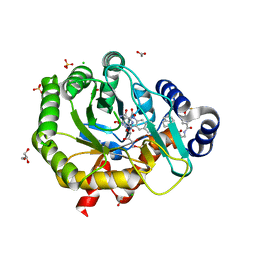 | | Crystal structure of human dihydroorotate dehydrogenase with ML390 | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Lewis, T.A, Sykes, D.B, Law, J.M, Munoz, B, Scadden, D.T, Rustiguel, J.K, Nonato, M.C, Schreiber, S.L. | | Deposit date: | 2016-05-31 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Development of ML390: A Human DHODH Inhibitor That Induces Differentiation in Acute Myeloid Leukemia.
ACS Med Chem Lett, 7, 2016
|
|
5HJN
 
 | |
5HJQ
 
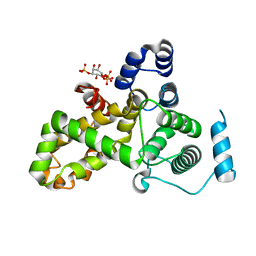 | | Crystal structure of the TBC domain of Skywalker/TBC1D24 from Drosophila melanogaster in complex with inositol(1,4,5)triphosphate | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, LD10117p | | Authors: | Fischer, B, Paesmans, J, Versees, W. | | Deposit date: | 2016-01-13 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Skywalker-TBC1D24 has a lipid-binding pocket mutated in epilepsy and required for synaptic function.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5KTG
 
 | |
5BTT
 
 | | Switching GFP fluorescence using genetically encoded phenyl azide chemistry through two different non-native post-translational modifications routes at the same position. | | Descriptor: | GLYCEROL, Green fluorescent protein, SULFATE ION | | Authors: | Hartley, A.M, Worthy, H.L, Reddington, S.C, Rizkallah, P.J, Jones, D.D. | | Deposit date: | 2015-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Molecular basis for functional switching of GFP by two disparate non-native post-translational modifications of a phenyl azide reaction handle.
Chem Sci, 7, 2016
|
|
5BT0
 
 | | Switching GFP fluorescence using genetically encoded phenyl azide chemistry through two different non-native post-translational modifications routes at the same position. | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | Hartley, A.M, Worthy, H.L, Reddington, S.C, Rizkallah, P.J, Jones, D.D. | | Deposit date: | 2015-06-02 | | Release date: | 2016-07-13 | | Last modified: | 2017-05-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for functional switching of GFP by two disparate non-native post-translational modifications of a phenyl azide reaction handle.
Chem Sci, 7, 2016
|
|
5DY6
 
 | | Enhanced superfolder GFP with DBCO at 148 | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | Jones, D.D, Rizkallah, P.J, Worthy, H.L. | | Deposit date: | 2015-09-24 | | Release date: | 2016-07-13 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Molecular basis for functional switching of GFP by two disparate non-native post-translational modifications of a phenyl azide reaction handle.
Chem Sci, 7, 2016
|
|
