8W2L
 
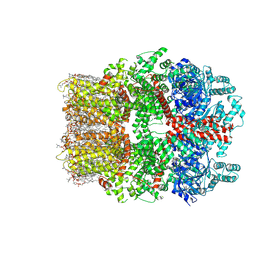 | | TRPM7 structure in complex with anticancer agent CCT128930 in closed state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 4-(4-chlorobenzyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-aminium, ... | | Authors: | Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2024-02-20 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural basis of selective TRPM7 inhibition by the anticancer agent CCT128930.
Cell Rep, 43, 2024
|
|
8Y0U
 
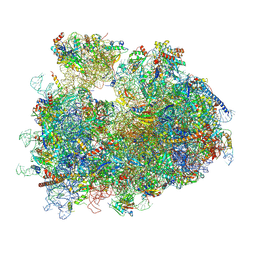 | | dormant ribosome with STM1 | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Du, M, Zeng, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | dormant ribosome with STM1
To Be Published
|
|
8Y0W
 
 | |
8Y0X
 
 | | Dormant ribosome with SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Du, M, Zeng, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The global structure of dormant ribosome
To Be Published
|
|
8XLD
 
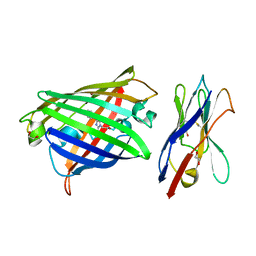 | | Structure of the GFP:GFP-nanobody complex from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Nanobody(Staygold-S2G10)-Nanobody(Staygold-S4F1), ZINC ION, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Bao, C. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the GFP:GFP-nanobody complex from Biortus.
To Be Published
|
|
8R6F
 
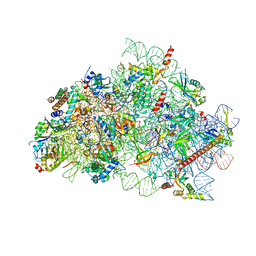 | | CryoEM structure of wheat 40S ribosomal subunit, body domain | | Descriptor: | 30S ribosomal protein S11, chloroplastic, 30S ribosomal protein S4, ... | | Authors: | Kravchenko, O.V, Baymukhametov, T.N, Afonina, Z.A, Vasilenko, K.S. | | Deposit date: | 2023-11-22 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | High-Resolution Structure and Internal Mobility of a Plant 40S Ribosomal Subunit.
Int J Mol Sci, 24, 2023
|
|
7GQU
 
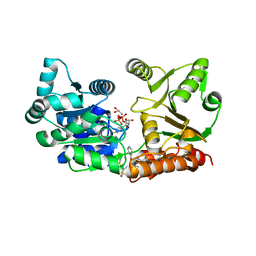 | | Crystal Structure of Werner helicase fragment 517-945 in covalent complex with N-[(E,1S)-1-cyclopropyl-3-methylsulfonylprop-2-enyl]-2-(1,1-difluoroethyl)-4-phenoxypyrimidine-5-carboxamide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, GLYCEROL, ... | | Authors: | Classen, M, Benz, J, Brugger, D, Tagliente, O, Rudolph, M.G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Chemoproteomic discovery of a covalent allosteric inhibitor of WRN helicase
Nature, 2024
|
|
7GQS
 
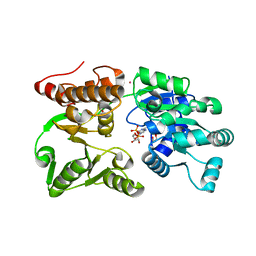 | | Crystal Structure of Werner helicase fragment 517-945 in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, ... | | Authors: | Classen, M, Benz, J, Brugger, D, Rudolph, M.G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Chemoproteomic discovery of a covalent allosteric inhibitor of WRN helicase
Nature, 2024
|
|
7GQT
 
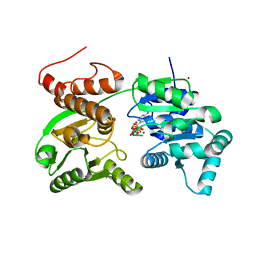 | | Crystal Structure of Werner helicase fragment 517-945 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, MAGNESIUM ION, ... | | Authors: | Classen, M, Benz, J, Brugger, D, Rudolph, M.G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Chemoproteomic discovery of a covalent allosteric inhibitor of WRN helicase
Nature, 2024
|
|
8UBG
 
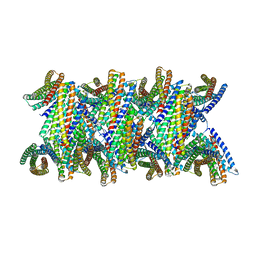 | | DpHF19 filament | | Descriptor: | DpHF19,Green fluorescent protein (Fragment) | | Authors: | Lynch, E.M, Shen, H, Kollman, J.M, Baker, D. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | De novo design of pH-responsive self-assembling helical protein filaments.
Nat Nanotechnol, 2024
|
|
8UB6
 
 | |
8U24
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 9 | | Descriptor: | Green Fluorescent Protein Variant #9, ccGFP 9 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U21
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP E6 | | Descriptor: | Green Fluorescent Protein Variant E6, ccGFP E6 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U23
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 8 | | Descriptor: | Green Fluorescent Protein Variant #8, ccGFP 8 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U20
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 5 | | Descriptor: | Green Fluorescent Protein Variant #5, ccGFP 5 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U22
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 7 | | Descriptor: | Green Fluorescent Protein Variant #7, ccGFP 7 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U1U
 
 | | Structure of a class A GPCR/agonist complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-C motif chemokine 1,C-C chemokine receptor type 8,EGFP fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Sun, D, Johnson, M, Masureel, M. | | Deposit date: | 2023-09-02 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
8TU9
 
 | | Cryo-EM structure of HGSNAT-acetyl-CoA complex at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, Enhanced green fluorescent protein,Heparan-alpha-glucosaminide N-acetyltransferase,Isoform 2 of Heparan-alpha-glucosaminide N-acetyltransferase | | Authors: | Navratna, V, Kumar, A, Mosalaganti, S. | | Deposit date: | 2023-08-15 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the human heparan-alpha-glucosaminide N-acetyltransferase (HGSNAT)
eLife, 13, 2024
|
|
7GFP
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with RAL-THA-6b94ceba-5 (Mpro-x11801) | | Descriptor: | 2-[3-(acetamidomethyl)-5-chlorophenyl]-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
8Q5I
 
 | | Structure of Candida albicans 80S ribosome in complex with cephaeline | | Descriptor: | 18S ribosomal RNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Atamas, A, Guskov, A, Yusupov, M. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural characterization of cephaeline binding to the eukaryotic ribosome using Cryo-Electron Microscopy
Biopolym Cell, 2023
|
|
8TLM
 
 | | Structure of a class A GPCR/Fab complex | | Descriptor: | C-C chemokine receptor type 8, Green fluorescent protein fusion, Fab heavy chain, ... | | Authors: | Sun, D, Johnson, M, Masureel, M. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
8THR
 
 | | Structure of the human vesicular monoamine transporter 2 (VMAT2) bound to tetrabenazine in an occluded conformation | | Descriptor: | (3S,5R,11bS)-9,10-dimethoxy-3-(2-methylpropyl)-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-2-one, fluorescent protein mVenus,Synaptic vesicular amine transporter,GFP nano body,Synaptic vesicular amine transporter,Synaptic vesicular amine transporter | | Authors: | Dalton, M.P, Coleman, J.A. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structure of the human vesicular monoamine transporter 2 (VMAT2) bound to tetrabenazine in an occluded conformation
To Be Published
|
|
8K2P
 
 | | Crystal structure of CtGST-F76A | | Descriptor: | Glutathione S-transferase | | Authors: | Yang, J, Xiao, J.Y, Lei, X.G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
8TGP
 
 | |
8PPK
 
 | | Bat-Hp-CoV Nsp1 and eIF1 bound to the human 40S small ribosomal subunit | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
