2RBQ
 
 | |
6BSK
 
 | | Human PIM1 kinase in complex with compound 12b | | Descriptor: | 1,2-ETHANEDIOL, 4-{6-[6-(propan-2-ylamino)-1H-indazol-1-yl]pyrazin-2-yl}benzoic acid, SULFATE ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2017-12-03 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.573 Å) | | Cite: | Discovery of 2,6-disubstituted pyrazine derivatives as inhibitors of CK2 and PIM kinases.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
7CSN
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 1.00 A resolution | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Viswanathan, V, Sharma, P, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-08-15 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 1.00 A resolution
To Be Published
|
|
6ETD
 
 | |
7CGP
 
 | | Cryo-EM structure of the human mitochondrial translocase TIM22 complex at 3.7 angstrom. | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Acylglycerol kinase, mitochondrial, ... | | Authors: | Qi, L, Wang, Q, Guan, Z, Yan, C, Yin, P. | | Deposit date: | 2020-07-01 | | Release date: | 2020-10-14 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the human mitochondrial translocase TIM22 complex.
Cell Res., 31, 2021
|
|
7SS8
 
 | | Human P300 complexed with a proline-based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-[1-(4-chlorophenyl)cyclopentane-1-carbonyl]-N-{[3-(methylcarbamoyl)phenyl]methyl}-D-prolinamide, Histone acetyltransferase p300, ... | | Authors: | Shewchuk, L.M, Reid, R.A. | | Deposit date: | 2021-11-10 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of Proline-Based p300/CBP Inhibitors Using DNA-Encoded Library Technology in Combination with High-Throughput Screening.
J.Med.Chem., 65, 2022
|
|
6H8V
 
 | | Beta-phosphoglucomutase from Lactococcus lactis in an open conformer in the P21 spacegroup to 1.8 A. | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, Beta-phosphoglucomutase, ... | | Authors: | Robertson, A.J, Bisson, C, Waltho, J.P. | | Deposit date: | 2018-08-03 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Transition state of phospho-enzyme hydrolysis in beta-phosphoglucomutase.
To Be Published
|
|
6H93
 
 | | Beta-phosphoglucomutase from Lactococcus lactis with inorganic phosphate bound in an open conformer to 1.8 A. | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ACETATE ION, ... | | Authors: | Robertson, A.J, Bisson, C, Waltho, J.P. | | Deposit date: | 2018-08-03 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Transition state of phospho-enzyme hydrolysis in beta-phosphoglucomutase.
To Be Published
|
|
5YT3
 
 | | Structure of the Human Mitogen-Activated Protein Kinase Kinase 1 S218D and S222D mutant | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase kinase 1, isoform CRA_d, ... | | Authors: | Nakae, S, Doko, K, Tada, T, Shirai, T. | | Deposit date: | 2017-11-16 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Human Mitogen-Activated Protein Kinase Kinase 1 S218D and S222D mutant
To Be Published
|
|
6HA6
 
 | | Factor Inhibiting HIF (FIH) in complex with zinc, NOG and TRPV3 (220-246) | | Descriptor: | GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, N-OXALYLGLYCINE, ... | | Authors: | Leissing, T.M, Clifton, I.J, Saward, B.G, Lu, X, Hopkinson, R.J, Schofield, C.J. | | Deposit date: | 2018-08-07 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Factor Inhibiting HIF (FIH) in complex with zinc, NOG and TRPV3 (220-246)
To Be Published
|
|
9F2H
 
 | | Crystal structure of SARS-CoV-2 N-protein C-terminal domain in complex with riluzole | | Descriptor: | 1,2-ETHANEDIOL, 6-(trifluoromethoxy)-1,3-benzothiazol-2-amine, Nucleoprotein, ... | | Authors: | Marquez-Monino, M.A, Gonzalez, B, Perez-Canadillas, J.M. | | Deposit date: | 2024-04-23 | | Release date: | 2024-11-13 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The ALS drug riluzole binds to the C-terminal domain of SARS-CoV-2 nucleocapsid protein and has antiviral activity.
Structure, 33, 2025
|
|
5GYG
 
 | |
6QL2
 
 | | Crystal structure of chimeric carbonic anhydrase VI with ethoxzolamide. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-ethoxy-1,3-benzothiazole-2-sulfonamide, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2019-01-31 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineered Carbonic Anhydrase VI-Mimic Enzyme Switched the Structure and Affinities of Inhibitors.
Sci Rep, 9, 2019
|
|
6BRE
 
 | | Hen Egg-White Lysozyme cocrystallized with Cadmium sulphate using CuKalpha source. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Sakon, J, Caviness, P, Jack, T, Pickens, J.B, Reed, P, Ruth, C. | | Deposit date: | 2017-11-30 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Hen Egg-White Lysozyme (HEWL) cocrystallized with Cadmium sulphate.
To Be Published
|
|
5K6X
 
 | | Sidekick-2 immunoglobulin domains 1-4, crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of sidekick-mediated cell-cell adhesion and specificity.
Elife, 5, 2016
|
|
8RRZ
 
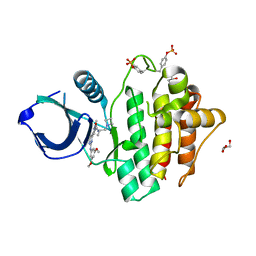 | | Crystal structure of SYK kinase in complex with compound 1 | | Descriptor: | GLYCEROL, N-[(2S)-1-(azetidin-1-yl)propan-2-yl]-3-{2-[(3,5-dimethoxyphenyl)amino]pyrimidin-4-yl}-1-methyl-1H-pyrazole-5-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Canevari, G. | | Deposit date: | 2024-01-24 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery and optimization of 4-pyrazolyl-2-aminopyrimidine derivatives as potent spleen tyrosine kinase (SYK) inhibitors.
Eur.J.Med.Chem., 270, 2024
|
|
6QLY
 
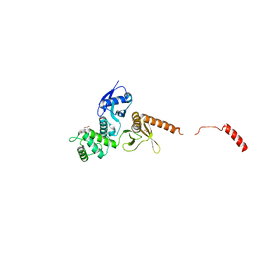 | | IDOL FERM domain | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase MYLIP, SULFATE ION | | Authors: | Martinelli, L, Sixma, T.K. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the LDL receptor-interacting FERM domain in the E3 ubiquitin ligase IDOL reveals an obscured substrate-binding site.
J.Biol.Chem., 295, 2020
|
|
9D0G
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with O-cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.50A resolution | | Descriptor: | (4S,5aS,8S,8aR)-4-(2-methylpropyl)-N-[(1S,5Z,7R,8R,9R,10R,11S,12R)-10,11,12-trihydroxy-7-methyl-2,13-dioxabicyclo[7.3.1]tridec-5-en-8-yl]octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Robinson, P.J, Benedetto, A.E, Yu, M, Tresco, B.I.C, See, D.N.Y, Jiang, T, Ramkissoon, A, Dunand, C.F, Svetlov, M.S, Lee, J, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Why Sulfur is Important in Lincosamide Antibiotics.
Chem, 11, 2025
|
|
6F4U
 
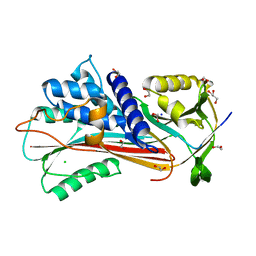 | |
5LZ4
 
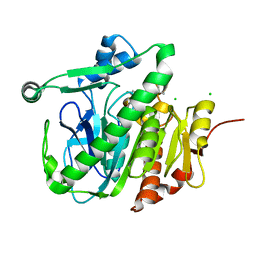 | | Fragment-based inhibitors of Lipoprotein associated Phospholipase A2 | | Descriptor: | 5-[2-(4,4-dimethyl-2-oxidanylidene-pyrrolidin-1-yl)ethoxy]-2-fluoranyl-benzenecarbonitrile, CHLORIDE ION, Platelet-activating factor acetylhydrolase | | Authors: | Woolford, A, Day, P. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Fragment-Based Approach to the Development of an Orally Bioavailable Lactam Inhibitor of Lipoprotein-Associated Phospholipase A2 (Lp-PLA2).
J. Med. Chem., 59, 2016
|
|
6HB6
 
 | | Crystal structure of E. coli tyrRS in complex with 5'-O-(N-L-tyrosyl)sulfamoyl-uridine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Tyrosine--tRNA ligase, ... | | Authors: | De Graef, S, Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2018-08-09 | | Release date: | 2019-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Comparative analysis of pyrimidine substituted aminoacyl-sulfamoyl nucleosides as potential inhibitors targeting class I aminoacyl-tRNA synthetases.
Eur.J.Med.Chem., 173, 2019
|
|
6Q4U
 
 | | KlenTaq DNA pol in a closed ternary complex with 7-deaza-7-(2-(2-hydroxyethoxy)-N-(prop-2-yn-1-yl)acetamide)-2-dATP | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*CP*TP*GP*TP*GP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*CP*AP*(DOC))-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | The Structure of an Archaeal B-Family DNA Polymerase in Complex with a Chemically Modified Nucleotide.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6BNI
 
 | |
6Q56
 
 | | Crystal structure of the B. subtilis M1A22 tRNA methyltransferase TrmK | | Descriptor: | NICKEL (II) ION, tRNA (adenine(22)-N(1))-methyltransferase | | Authors: | Degut, C, Roovers, M, Barraud, P, Brachet, F, Feller, A, Larue, V, Al Refaii, A, Caillet, J, Droogmans, L, Tisne, C. | | Deposit date: | 2018-12-07 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of B. subtilis m1A22 tRNA methyltransferase TrmK: insights into tRNA recognition.
Nucleic Acids Res., 47, 2019
|
|
6HHO
 
 | | Crystal structure of RIP1 kinase in complex with GSK547 | | Descriptor: | 6-[4-[(5~{S})-5-[3,5-bis(fluoranyl)phenyl]pyrazolidin-1-yl]carbonylpiperidin-1-yl]pyrimidine-4-carbonitrile, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Thorpe, J.H, Harris, P.A. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | RIP1 Kinase Drives Macrophage-Mediated Adaptive Immune Tolerance in Pancreatic Cancer.
Cancer Cell, 34, 2018
|
|
