6YCD
 
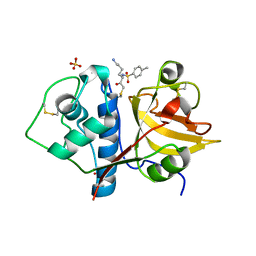 | | Structure the ananain protease from Ananas comosus covalently bound to the TLCK inhibitor | | Descriptor: | Ananain, GLYCEROL, N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
6YCG
 
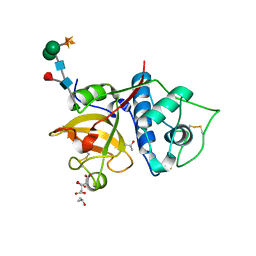 | | Structure the bromelain protease from Ananas comosus in complex with the TLCK inhibitor | | Descriptor: | CITRIC ACID, FBSB, ISOPROPYL ALCOHOL, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
6YCC
 
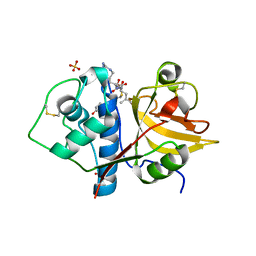 | | Structure the ananain protease from Ananas comosus covalently bound to the E64 inhibitor | | Descriptor: | Ananain, GLYCEROL, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
7KM4
 
 | |
5XZG
 
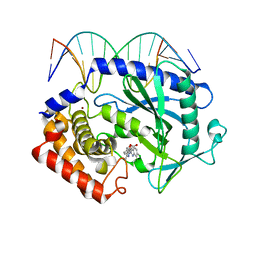 | | Mouse cGAS bound to the inhibitor RU521 | | Descriptor: | 2-(4,5-dichloro-1H-benzimidazol-2-yl)-5-methyl-4-[(1R)-3-oxo-1,3-dihydro-2-benzofuran-1-yl]-1,2-dihydro-3H-pyrazol-3-one, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*G)-3'), ... | | Authors: | Vincent, J, Adura, C, Gao, P, Luz, A, Lama, L, Asano, Y, Okamoto, R, Imaeda, T, Aida, J, Rothamel, K, Gogakos, T, Steinberg, J, Reasoner, S, Aso, K, Tuschl, T, Patel, D.J, Glickman, J.F, Ascano, M. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice.
Nat Commun, 8, 2017
|
|
7W82
 
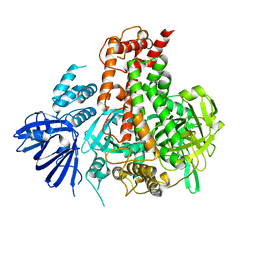 | | Crystal structure of maize RDR2 | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Du, X, Yang, Z, Du, J. | | Deposit date: | 2021-12-07 | | Release date: | 2022-06-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of plant RNA-DEPENDENT RNA POLYMERASE 2, an enzyme involved in small interfering RNA production.
Plant Cell, 34, 2022
|
|
7W88
 
 | |
7W84
 
 | |
3L8E
 
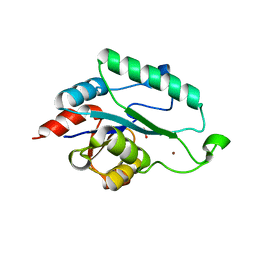 | | Crystal Structure of apo form of D,D-heptose 1.7-bisphosphate phosphatase from E. Coli | | Descriptor: | ACETIC ACID, D,D-heptose 1,7-bisphosphate phosphatase, ZINC ION | | Authors: | Nguyen, H, Peisach, E, Allen, K.N. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Determinants of Substrate Recognition in the HAD Superfamily Member d-glycero-d-manno-Heptose-1,7-bisphosphate Phosphatase (GmhB) .
Biochemistry, 49, 2010
|
|
3GPM
 
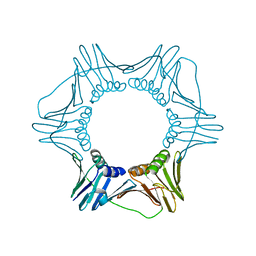 | | Structure of the trimeric form of the E113G PCNA mutant protein | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Freudenthal, B.D, Gakhar, L, Ramaswamy, S, Washington, M.T. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A charged residue at the subunit interface of PCNA promotes trimer formation by destabilizing alternate subunit interactions.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5XZB
 
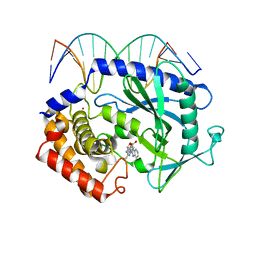 | | Mouse cGAS bound to the inhibitor RU365 | | Descriptor: | (3R)-3-[1-(1H-benzimidazol-2-yl)-5-hydroxy-3-methyl-1H-pyrazol-4-yl]-2-benzofuran-1(3H)-one, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*G)-3'), ... | | Authors: | Vincent, J, Adura, C, Gao, P, Luz, A, Lama, L, Asano, Y, Okamoto, R, Imaeda, T, Aida, J, Rothamel, K, Gogakos, T, Steinberg, J, Reasoner, S, Aso, K, Tuschl, T, Patel, D.J, Glickman, J.F, Ascano, M. | | Deposit date: | 2017-07-12 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice.
Nat Commun, 8, 2017
|
|
6H35
 
 | | Myxococcus xanthus MglA bound to GDP and Pi with mixed inactive and active switch region conformations | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GUANOSINE-5'-DIPHOSPHATE, Mutual gliding-motility protein MglA, ... | | Authors: | Varela, P.F, Galicia, C, Cherfils, J. | | Deposit date: | 2018-07-17 | | Release date: | 2019-12-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | MglA functions as a three-state GTPase to control movement reversals of Myxococcus xanthus.
Nat Commun, 10, 2019
|
|
3L8F
 
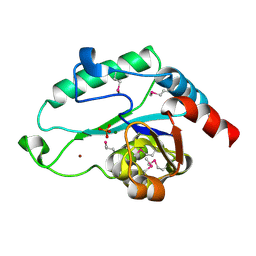 | | Crystal Structure of D,D-heptose 1.7-bisphosphate phosphatase from E. Coli complexed with magnesium and phosphate | | Descriptor: | D,D-heptose 1,7-bisphosphate phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Nguyen, H, Peisach, E, Allen, K.N. | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Determinants of Substrate Recognition in the HAD Superfamily Member d-glycero-d-manno-Heptose-1,7-bisphosphate Phosphatase (GmhB) .
Biochemistry, 49, 2010
|
|
5HGR
 
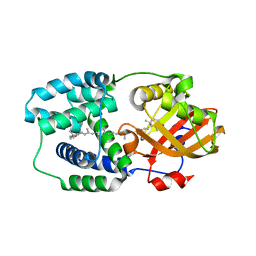 | | Structure of Anabaena (Nostoc) sp. PCC 7120 Orange Carotenoid Protein binding canthaxanthin | | Descriptor: | Orange Carotenoid Protein (OCP), beta,beta-carotene-4,4'-dione | | Authors: | Sutter, M, Leverenz, R.L, Kerfeld, C.A. | | Deposit date: | 2016-01-08 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.681 Å) | | Cite: | Different Functions of the Paralogs to the N-Terminal Domain of the Orange Carotenoid Protein in the Cyanobacterium Anabaena sp. PCC 7120.
Plant Physiol., 171, 2016
|
|
5Z8S
 
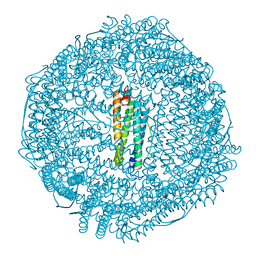 | |
5Z91
 
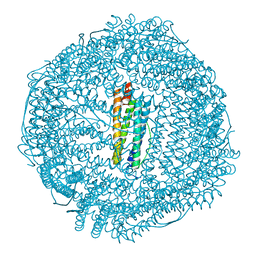 | | Human mitochondrial ferritin mutant bound with gold ions | | Descriptor: | Ferritin, mitochondrial, GOLD ION, ... | | Authors: | Zang, J, Zheng, B, Zhao, G. | | Deposit date: | 2018-02-01 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Design and site-directed compartmentalization of gold nanoclusters within the intrasubunit interfaces of ferritin nanocage.
J Nanobiotechnology, 17, 2019
|
|
5Z8U
 
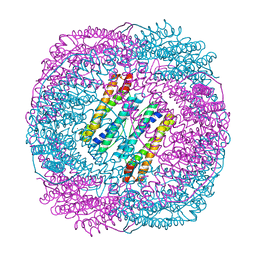 | | Human mitochondrial ferritin mutant - C102A/C130A | | Descriptor: | Ferritin, mitochondrial, MAGNESIUM ION | | Authors: | Zang, J, Zheng, B, Zhao, G. | | Deposit date: | 2018-02-01 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and site-directed compartmentalization of gold nanoclusters within the intrasubunit interfaces of ferritin nanocage.
J Nanobiotechnology, 17, 2019
|
|
6H5B
 
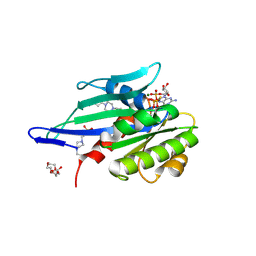 | | Myxococcus xanthus MglA in complex with its GAP MglB and GTPgammaS | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Galicia, C, Cherfils, J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-11-27 | | Last modified: | 2020-11-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | MglA functions as a three-state GTPase to control movement reversals of Myxococcus xanthus.
Nat Commun, 10, 2019
|
|
5Z8J
 
 | |
6HJH
 
 | | Myxococcus xanthus MglA bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Mutual gliding-motility protein MglA | | Authors: | Varela, P.F, Cherfils, J. | | Deposit date: | 2018-09-03 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | MglA functions as a three-state GTPase to control movement reversals of Myxococcus xanthus.
Nat Commun, 10, 2019
|
|
5KV9
 
 | | Crystal structure of a hPIV haemagglutinin-neuraminidase-inhibitor complex | | Descriptor: | 1,2-ETHANEDIOL, 2,6-anhydro-3,4,5-trideoxy-5-[(2-methylpropanoyl)amino]-4-(4-phenyl-1H-1,2,3-triazol-1-yl)-D-glycero-D-galacto-non-2-en onic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dirr, L, El-Deeb, I.M, Chavas, L.M.G, Guillon, P, von Itzstein, M. | | Deposit date: | 2016-07-13 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The impact of the butterfly effect on human parainfluenza virus haemagglutinin-neuraminidase inhibitor design.
Sci Rep, 7, 2017
|
|
8SLQ
 
 | | Cryo-EM structure of the rat TRPM5 channel in 2mM calcium, high-3 | | Descriptor: | Transient receptor potential cation channel subfamily M member 5 | | Authors: | Karuppan, S, Schrag, L.G, Jara-Oseguera, A, Zubcevic, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural dynamics at cytosolic interprotomer interfaces control gating of a mammalian TRPM5 channel.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SLE
 
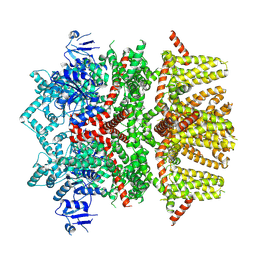 | | Cryo-EM structure of the rat TRPM5 channel in trace calcium, trace-3 | | Descriptor: | Transient receptor potential cation channel subfamily M member 5 | | Authors: | Karuppan, S, Schrag, L.G, Jara-Oseguera, A, Zubcevic, L. | | Deposit date: | 2023-04-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural dynamics at cytosolic interprotomer interfaces control gating of a mammalian TRPM5 channel.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SLI
 
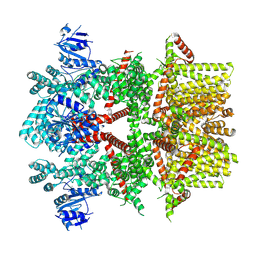 | | Cryo-EM structure of the rat TRPM5 channel in 2mM calcium, high-1 | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily M member 5 | | Authors: | Karuppan, S, Schrag, L.G, Jara-Oseguera, A, Zubcevic, L. | | Deposit date: | 2023-04-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural dynamics at cytosolic interprotomer interfaces control gating of a mammalian TRPM5 channel.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7LHQ
 
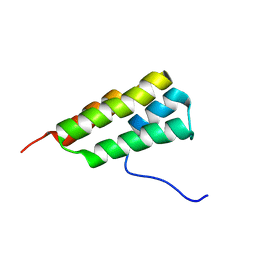 | | Solution structure of SARS-CoV-2 nonstructural protein 7 at pH 7.0 | | Descriptor: | Non-structural protein 7 | | Authors: | Lee, Y, Tonelli, M, Anderson, T.K, Kirchdoerfer, R.N, Henzler-Wildman, K, Lee, W. | | Deposit date: | 2021-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | pH-dependent polymorphism of the structure of SARS-CoV-2 nsp7
Biorxiv, 2021
|
|
