4YM6
 
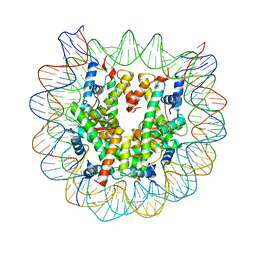 | | Crystal structure of the human nucleosome containing 6-4PP (outside) | | Descriptor: | 145-MER DNA, Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.514 Å) | | Cite: | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
6HGN
 
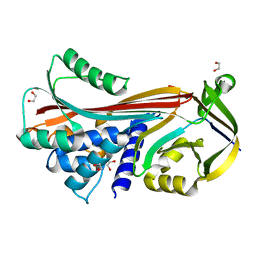 | |
6SUA
 
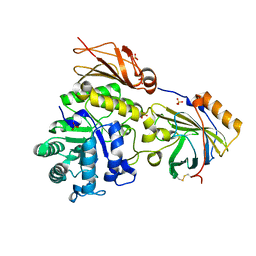 | |
6SNU
 
 | | Crystal structure of the W60C mutant of the (S)-selective transaminase from Chromobacterium violaceum | | Descriptor: | 1,2-ETHANEDIOL, Aspartate aminotransferase family protein, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ruggieri, F, Gustafsson, C, Kimbung, R.Y, Walse, B, Logan, D.T, Berglund, P. | | Deposit date: | 2019-08-27 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Combined with Molecular Dynamics Reveal Altered Flow of Water in the Active Site of W60C Chromobacterium violaceum omega-transaminase
Not Published
|
|
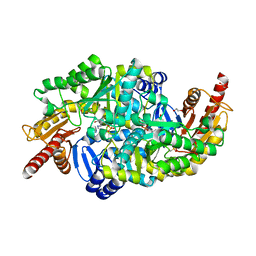
4GPW
 
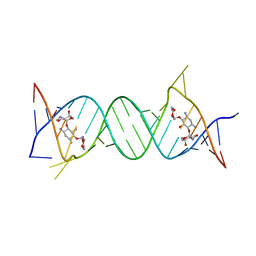 | | Crystal structure of the protozoal cytoplasmic ribosomal decoding site in complex with 6'-hydroxysisomicin (P21212 form) | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(hydroxymethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Koganei, M, Maianti, J.P, Ly, V.L, Hanessian, S. | | Deposit date: | 2012-08-22 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of a bioactive 6'-hydroxy variant of sisomicin bound to the bacterial and protozoal ribosomal decoding sites
Chemmedchem, 8, 2013
|
|
4GPY
 
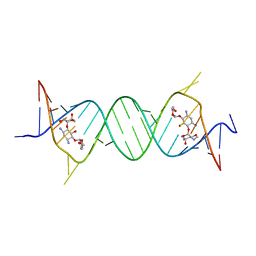 | | Crystal structure of the bacterial ribosomal decoding site in complex with 6'-hydroxysisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(hydroxymethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Koganei, M, Maianti, J.P, Ly, V.L, Hanessian, S. | | Deposit date: | 2012-08-22 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of a bioactive 6'-hydroxy variant of sisomicin bound to the bacterial and protozoal ribosomal decoding sites
Chemmedchem, 8, 2013
|
|
4F8V
 
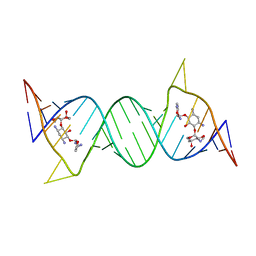 | | Crystal structure of the bacterial ribosomal decoding site in complex with sisomicin (P21212 form) | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, RNA (5'-R(P*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Koganei, M, Kasahara, T. | | Deposit date: | 2012-05-18 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and specific binding mode of sisomicin to the bacterial ribosomal decoding site.
Acs Med.Chem.Lett., 3, 2012
|
|
6T3B
 
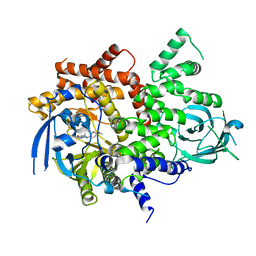 | | Crystal structure of PI3Kgamma with a dihydropurinone inhibitor (compound 4) | | Descriptor: | 2-[(4-methoxy-2-methyl-phenyl)amino]-7-methyl-9-(4-oxidanylcyclohexyl)purin-8-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Petersen, J, Oster, L, Schimpl, M, Goldberg, F.W, Finlay, M.R.V, Ting, A.K.T, Beattie, D, Lamont, G.M, Fallan, C, Wrigley, G.L, Howard, M.R, Williamson, B, Davies, B.R, Cadogan, E.B, Ramos-Montoya, A, Dean, E. | | Deposit date: | 2019-10-10 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | The Discovery of 7-Methyl-2-[(7-methyl[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8-one (AZD7648), a Potent and Selective DNA-Dependent Protein Kinase (DNA-PK) Inhibitor.
J.Med.Chem., 63, 2020
|
|
4YDQ
 
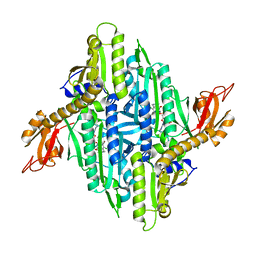 | | Crystal Structure of Prolyl-tRNA Synthetase (PRS) from Plasmodium falciparum in complex with Halofuginone and AMPPNP | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Jain, V, Yogavel, M, Sharma, A. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Prolyl-tRNA Synthetase-Halofuginone Complex Provides Basis for Development of Drugs against Malaria and Toxoplasmosis
Structure, 23, 2015
|
|
6IC6
 
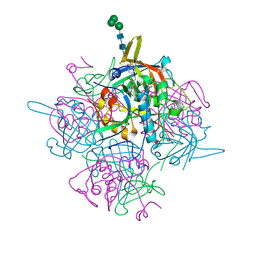 | | Human cathepsin-C in complex with cyclopropyl peptidyl nitrile inhibitor 1 | | Descriptor: | (2~{S})-~{N}-[(1~{R},2~{R})-1-(aminomethyl)-2-[4-[4-(trifluoromethyl)phenyl]phenyl]cyclopropyl]-2-azanyl-butanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakansson, M, Logan, D.T, Korkmaz, B, Lesner, A, Wysocka, M, Gieldon, A, Gauthier, F, Jenne, D, Lauritzen, C, Pedersen, J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Structure-based design and in vivo anti-arthritic activity evaluation of a potent dipeptidyl cyclopropyl nitrile inhibitor of cathepsin C.
Biochem. Pharmacol., 164, 2019
|
|
6SKX
 
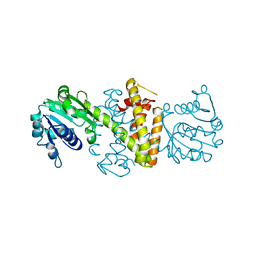 | | Structure of Reductive Aminase from Neosartorya fumigata | | Descriptor: | Oxidoreductase, putative | | Authors: | Sharma, M, Mangas-Sanchez, J, Turner, N.J, Grogan, G. | | Deposit date: | 2019-08-16 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Asymmetric synthesis of primary amines catalyzed by thermotolerant fungal reductive aminases.
Chem Sci, 11, 2020
|
|
2OGW
 
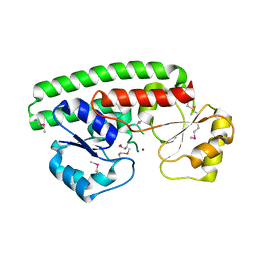 | | Structure of ABC type zinc transporter from E. coli | | Descriptor: | High-affinity zinc uptake system protein znuA precursor, ZINC ION | | Authors: | Sharma, A, Yogavel, M. | | Deposit date: | 2007-01-09 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Analysis of ABC-family Periplasmic Zinc Binding Protein Provides New Insights Into Mechanism of Ligand Uptake and Release
J.Mol.Biol., 367, 2007
|
|
6ICG
 
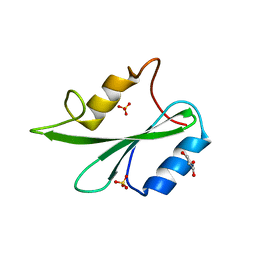 | | Grb2 SH2 domain in phosphopeptide free form | | Descriptor: | GLYCEROL, Growth factor receptor-bound protein 2, SULFATE ION | | Authors: | Hosoe, Y, Numoto, N, Inaba, S, Ogawa, S, Morii, H, Abe, R, Ito, N, Oda, M. | | Deposit date: | 2018-09-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural and functional properties of Grb2 SH2 dimer in CD28 binding.
Biophys Physicobio., 16, 2019
|
|
4YM5
 
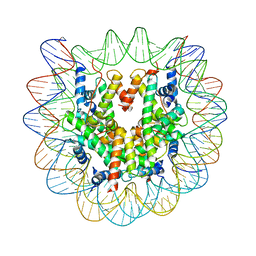 | | Crystal structure of the human nucleosome containing 6-4PP (inside) | | Descriptor: | 144 mer-DNA, 144-mer DNA, Histone H2A type 1-B/E, ... | | Authors: | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.005 Å) | | Cite: | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
6TF7
 
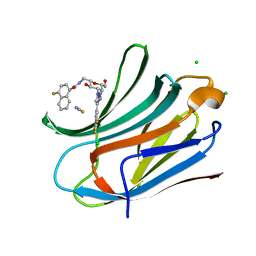 | | Human galectin-3c in complex with a galactose derivative | | Descriptor: | 4-fluoranyl-~{N}-[[(2~{S},3~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-2-yl]methyl]naphthalene-1-carboxamide, CHLORIDE ION, Galectin-3, ... | | Authors: | Nilsson, U.J, Zetterberg, F, Hakansson, M, Logan, D.T. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 3-Substituted 1-Naphthamidomethyl-C-galactosyls Interact with Two Unique Sub-sites for High-Affinity and High-Selectivity Inhibition of Galectin-3.
Molecules, 24, 2019
|
|
4WJV
 
 | | Crystal structure of Rsa4 in complex with the Nsa2 binding peptide | | Descriptor: | Maltose-binding periplasmic protein, Ribosome assembly protein 4, Ribosome biogenesis protein NSA2, ... | | Authors: | Holdermann, I, Paternoga, H, Bassler, J, Hurt, E, Sinning, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
6T3C
 
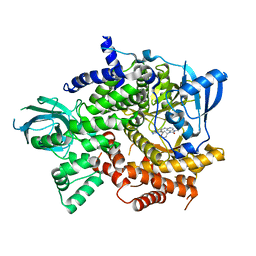 | | Crystal structure of PI3Kgamma in complex with DNA-PK inhibitor AZD7648 | | Descriptor: | 7-methyl-2-[(7-methyl-[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(oxan-4-yl)purin-8-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Schimpl, M, Goldberg, F.W, Finlay, M.R.V, Ting, A.K.T, Beattie, D, Lamont, G.M, Fallan, C, Wrigley, G.L, Howard, M.R, Williamson, B, Davies, B.R, Cadogan, E.B, Ramos-Montoya, A, Dean, E. | | Deposit date: | 2019-10-10 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The Discovery of 7-Methyl-2-[(7-methyl[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8-one (AZD7648), a Potent and Selective DNA-Dependent Protein Kinase (DNA-PK) Inhibitor.
J.Med.Chem., 63, 2020
|
|
6RA4
 
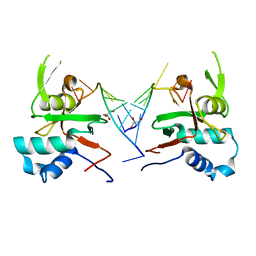 | | Human ARGONAUTE-2 PAZ DOMAIN (214-347) IN COMPLEX WITH CGUGACUCU | | Descriptor: | GLYCEROL, Protein argonaute-2, RNA (5'-R(*CP*GP*UP*GP*AP*CP*UP*CP*U)-3') | | Authors: | Rondeau, J.-M, Bourgier, E. | | Deposit date: | 2019-04-05 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How to Computationally Stack the Deck for Hit-to-Lead Generation: In Silico Molecular Interaction Energy Profiling for de Novo siRNA Guide Strand Surrogate Selection.
J.Chem.Inf.Model., 59, 2019
|
|
6HGI
 
 | |
4YCN
 
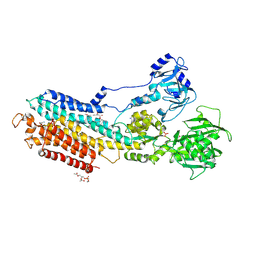 | | Crystal structure of the calcium pump with bound marine macrolide BLLB | | Descriptor: | (4S,5E,8S,9E,11S,13E,15E,18R)-4-hydroxy-8-methoxy-9,11-dimethyl-18-[(1Z,4E)-2-methylhexa-1,4-dien-1-yl]oxacyclooctadeca-5,9,13,15-tetraen-2-one, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Morita, M, Ogawa, H, Ohno, O, Yamori, T, Suenaga, K, Toyoshima, C. | | Deposit date: | 2015-02-20 | | Release date: | 2016-01-13 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Biselyngbyasides, cytotoxic marine macrolides, are novel and potent inhibitors of the Ca(2+) pumps with a unique mode of binding
Febs Lett., 589, 2015
|
|
2P0I
 
 | | Crystal structure of L-rhamnonate dehydratase from Gibberella zeae | | Descriptor: | GLYCEROL, L-rhamnonate dehydratase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Dickey, M, Logan, C, Gheyi, T, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-13 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of L-Rhamnonate Dehydratase from Gibberella Zeae
To be Published
|
|
4P3S
 
 | |
4P3T
 
 | |
6I6S
 
 | | Circular permutant of ribosomal protein S6, adding 9aa to C terminal of P68-69, L75A mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6, POTASSIUM ION, SODIUM ION | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6IC7
 
 | | Human cathepsin-C in complex with dipeptidyl cyclopropyl nitrile inhibitor 3 | | Descriptor: | 1-azanyl-~{N}-[(1~{R},2~{R})-1-cyano-2-[4-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]phenyl]cyclopropyl]cyclohexane-1-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakansson, M, Logan, D.T, Korkmaz, B, Lesner, A, Wysocka, M, Gieldon, A, Gauthier, F, Jenne, D, Lauritzen, C, Pedersen, J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design and in vivo anti-arthritic activity evaluation of a potent dipeptidyl cyclopropyl nitrile inhibitor of cathepsin C.
Biochem. Pharmacol., 164, 2019
|
|
