9AT1
 
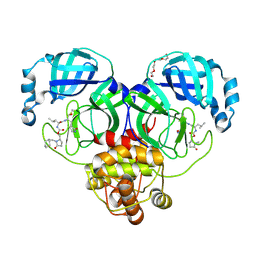 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a methylcyclohexyl 2-pyrrolidone inhibitor (R-enantiomer) | | Descriptor: | (1S,2S)-2-{[N-({[(2R)-1-(cyclohexylmethyl)-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9ASZ
 
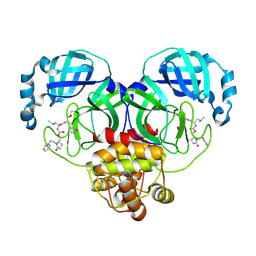 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a phenylethyl 2-pyrrolidone inhibitor | | Descriptor: | (1S,2S)-1-hydroxy-2-{[N-({[(2S)-5-oxo-1-(2-phenylethyl)pyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9ATF
 
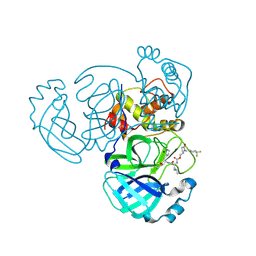 | | Crystal structure of MERS 3CL protease in complex with a 1-methyl-4,4-difluorocyclohexyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(4,4-difluorocyclohexyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(4,4-difluorocyclohexyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
2GSC
 
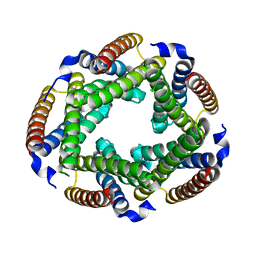 | | Crystal Structure of the Conserved Hypothetical Cytosolic Protein Xcc0516 from Xanthomonas campestris | | Descriptor: | Putative uncharacterized protein XCC0516 | | Authors: | Lin, L.Y, Ching, C.L, Chin, K.H, Chou, S.H, Chan, N.L. | | Deposit date: | 2006-04-26 | | Release date: | 2006-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the conserved hypothetical cytosolic protein Xcc0516 from Xanthomonas campestris reveals a novel quaternary structure assembled by five four-helix bundles.
Proteins, 65, 2006
|
|
4TNH
 
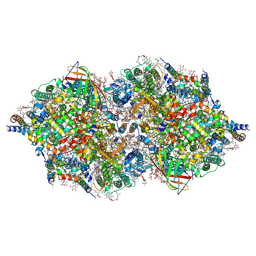 | | RT XFEL structure of Photosystem II in the dark state at 4.9 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.900007 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
2D27
 
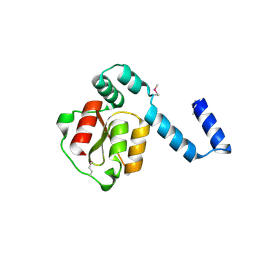 | | Structure of the N-terminal domain of XpsE (crystal form I4122) | | Descriptor: | type II secretion ATPase XpsE | | Authors: | Chen, Y, Shiue, S.-J, Huang, C.-W, Chang, J.-L, Chien, Y.-L, Hu, N.-T, Chan, N.-L. | | Deposit date: | 2005-09-03 | | Release date: | 2005-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure and Function of the XpsE N-Terminal Domain, an Essential Component of the Xanthomonas campestris Type II Secretion System
J.Biol.Chem., 280, 2005
|
|
5KL1
 
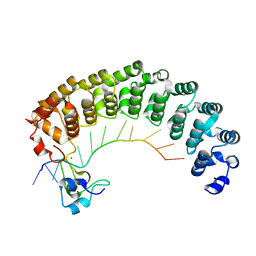 | | Crystal structure of the Pumilio-Nos-hunchback RNA complex | | Descriptor: | Maternal protein pumilio, Protein nanos, RNA (5'-R(*AP*AP*AP*UP*UP*GP*UP*AP*CP*AP*UP*A)-3'), ... | | Authors: | Qiu, C, Hall, T.M.T. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.701 Å) | | Cite: | Drosophila Nanos acts as a molecular clamp that modulates the RNA-binding and repression activities of Pumilio.
Elife, 5, 2016
|
|
5KL8
 
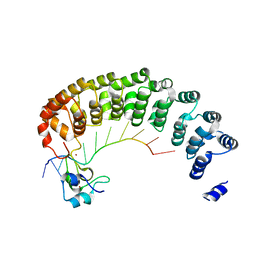 | | Crystal structure of the Pumilio-Nos-CyclinB RNA complex | | Descriptor: | Maternal protein pumilio, Protein nanos, RNA (5'-R(*UP*AP*UP*UP*UP*GP*UP*AP*AP*UP*U)-3'), ... | | Authors: | Qiu, C, Hall, T.M.T. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Drosophila Nanos acts as a molecular clamp that modulates the RNA-binding and repression activities of Pumilio.
Elife, 5, 2016
|
|
1VVC
 
 | |
2XBN
 
 | | Inhibition of the PLP-dependent enzyme serine palmitoyltransferase by cycloserine: evidence for a novel decarboxylative mechanism of inactivation | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, MAGNESIUM ION, SERINE PALMITOYLTRANSFERASE | | Authors: | Lowther, J, Yard, B.A, Johnson, K.A, Carter, L.G, Bhat, V.T, Raman, M.C.C, Clarke, D.J, Ramakers, B, McMahon, S.A, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2010-04-13 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of the Plp-Dependent Enzyme Serine Palmitoyltransferase by Cycloserine: Evidence for a Novel Decarboxylative Mechanism of Inactivation.
Mol.Biosystems, 6, 2010
|
|
7R3V
 
 | | Crystal structure of bovine Cytochrome bc1 in complex with inhibitor CK-2-67. | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Pinthong, N, Amporndanai, K, O'Neill, P.M, Hasnain, S.S, Antonyuk, S. | | Deposit date: | 2022-02-07 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Targeting the Ubiquinol-Reduction (Q i ) Site of the Mitochondrial Cytochrome bc 1 Complex for the Development of Next Generation Quinolone Antimalarials.
Biology (Basel), 11, 2022
|
|
5KLA
 
 | |
2G5G
 
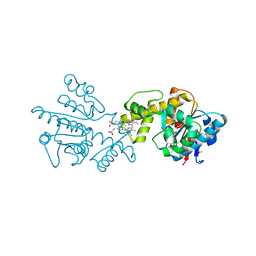 | |
1VVD
 
 | |
1VVE
 
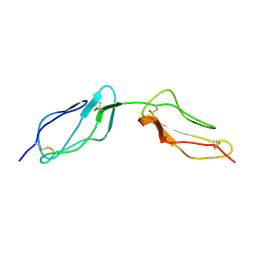 | |
5UHG
 
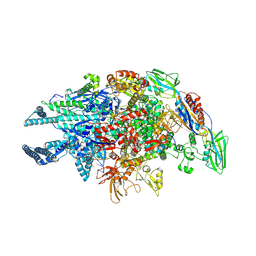 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex in complex with D-AAP1 and Rifampin | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*AP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.971 Å) | | Cite: | Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.
Mol. Cell, 66, 2017
|
|
6P3H
 
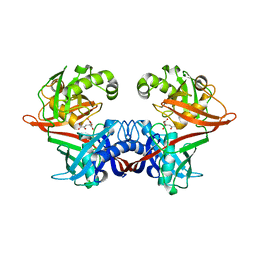 | | Crystal structure of LigU(K66M) bound to substrate | | Descriptor: | (1E)-4-oxobut-1-ene-1,2,4-tricarboxylic acid, (4E)-oxalomesaconate Delta-isomerase, CHLORIDE ION | | Authors: | Cory, S.A, Hogancamp, T.N, Raushel, F.M, Barondeau, D.P. | | Deposit date: | 2019-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure and Chemical Reaction Mechanism of LigU, an Enzyme That Catalyzes an Allylic Isomerization in the Bacterial Degradation of Lignin.
Biochemistry, 58, 2019
|
|
5UH6
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex containing 2ntRNA in complex with Rifampin | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*AP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.837 Å) | | Cite: | Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.
Mol. Cell, 66, 2017
|
|
5UHD
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex containing 4nt RNA in complex with Rifampin | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.01 Å) | | Cite: | Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.
Mol. Cell, 66, 2017
|
|
5UHC
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex containing 3nt RNA in complex with Rifampin | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*AP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.796 Å) | | Cite: | Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.
Mol. Cell, 66, 2017
|
|
6P3J
 
 | | Crystal structure of LigU | | Descriptor: | (4E)-oxalomesaconate Delta-isomerase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Cory, S.A, Hogancamp, T.N, Raushel, F.M, Barondeau, D.P. | | Deposit date: | 2019-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure and Chemical Reaction Mechanism of LigU, an Enzyme That Catalyzes an Allylic Isomerization in the Bacterial Degradation of Lignin.
Biochemistry, 58, 2019
|
|
7SH2
 
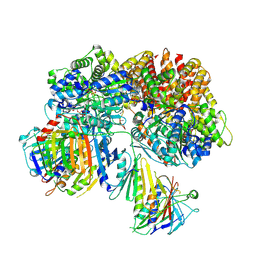 | | Structure of the yeast Rad24-RFC loader bound to DNA and the open 9-1-1 clamp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, Crick strand, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2021-10-07 | | Release date: | 2022-03-23 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | DNA is loaded through the 9-1-1 DNA checkpoint clamp in the opposite direction of the PCNA clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SGZ
 
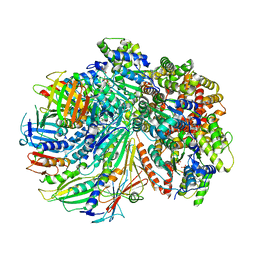 | | Structure of the yeast Rad24-RFC loader bound to DNA and the closed 9-1-1 clamp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, Crick strand, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2021-10-07 | | Release date: | 2022-03-23 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | DNA is loaded through the 9-1-1 DNA checkpoint clamp in the opposite direction of the PCNA clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1H4B
 
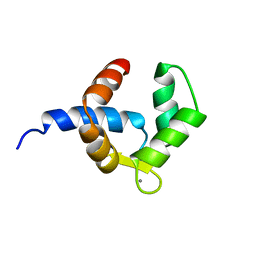 | | SOLUTION STRUCTURE OF THE BIRCH POLLEN ALLERGEN BET V 4 | | Descriptor: | CALCIUM ION, POLCALCIN BET V 4 | | Authors: | Neudecker, P, Nerkamp, J, Eisenmann, A, Lauber, T, Lehmann, K, Schweimer, K, Roesch, P. | | Deposit date: | 2003-02-26 | | Release date: | 2004-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics, and Hydrodynamics of the Calcium-Bound Cross-Reactive Birch Pollen Allergen Bet V 4 Reveal a Canonical Monomeric Two EF-Hand Assembly with a Regulatory Function
J.Mol.Biol., 336, 2004
|
|
2Y92
 
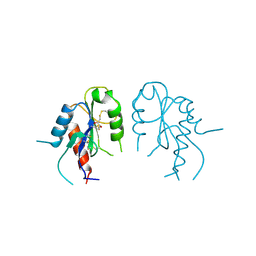 | | Crystal structure of MAL adaptor protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, TOLL/INTERLEUKIN-1 RECEPTOR DOMAIN-CONTAINING ADAPTER PROTEIN, | | Authors: | Valkov, E, Stamp, A, Martin, J.L, Kobe, B. | | Deposit date: | 2011-02-11 | | Release date: | 2011-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structure of Toll-Like Receptor Adaptor Mal/Tirap Reveals the Molecular Basis for Signal Transduction and Disease Protection.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
