2M34
 
 | |
7ZUN
 
 | | Crystal structure of PIM1 in complex with a Pyrrolo-Pyrazinone compound | | Descriptor: | (4~{S})-4-(2-azanylethyl)-6-phenyl-7-[3-(trifluoromethyloxy)phenyl]-3,4-dihydropyrrolo[1,2-a]pyrazin-1-ol, Isoform 2 of Serine/threonine-protein kinase pim-1 | | Authors: | Casale, E. | | Deposit date: | 2022-05-12 | | Release date: | 2022-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stereoselective synthesis of 3,4-dihydropyrrolo[1,2-a]pyrazin-1(2H)-one derivatives as PIM kinase inhibitors inspired from marine alkaloids.
Chirality, 34, 2022
|
|
3LWF
 
 | |
3CO5
 
 | | Crystal structure of sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae | | Descriptor: | BETA-MERCAPTOETHANOL, Putative two-component system transcriptional response regulator | | Authors: | Osipiuk, J, Hendricks, R, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of Sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae.
To be Published
|
|
3CLK
 
 | |
5KYJ
 
 | | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core | | Descriptor: | (6~{R})-5-(5-fluoranyl-2-methoxy-pyrimidin-4-yl)-2-(3-methylsulfonylphenyl)-6-propan-2-yl-4,6-dihydropyrrolo[3,4-c]pyrazole, Oxysterols receptor LXR-beta, Retinoic acid receptor RXR-beta | | Authors: | Chen, G, McKeever, B.M. | | Deposit date: | 2016-07-21 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
7W07
 
 | | Itaconate inducible LysR-Type Transcriptional regulator (ITCR) in complex with itaconate, Space group C121. | | Descriptor: | 2-methylidenebutanedioic acid, SULFATE ION, Transcriptional regulator, ... | | Authors: | Sun, P.K, Wang, B, Li, X.J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | A genetically encoded fluorescent biosensor for detecting itaconate with subcellular resolution in living macrophages.
Nat Commun, 13, 2022
|
|
7W08
 
 | |
7W06
 
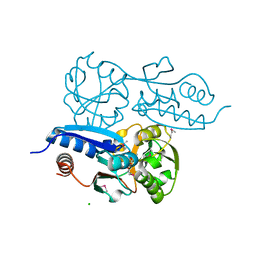 | | Itaconate inducible LysR-Type Transcriptional regulator (ITCR) in complex with itaconate (SeMet labeled), Space group C121. | | Descriptor: | 2-methylidenebutanedioic acid, CHLORIDE ION, SULFATE ION, ... | | Authors: | Sun, P.K, Wang, B, Wang, Z.X, Qi, S, Li, X.J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A genetically encoded fluorescent biosensor for detecting itaconate with subcellular resolution in living macrophages.
Nat Commun, 13, 2022
|
|
3CS3
 
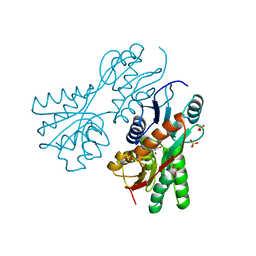 | | Crystal structure of sugar-binding transcriptional regulator (LacI family) from Enterococcus faecalis | | Descriptor: | GLYCEROL, SULFATE ION, Sugar-binding transcriptional regulator, ... | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Iizuka, M, Groshong, C, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-08 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of sugar-binding transcriptional regulator (LacI family) from Enterococcus faecalis.
To be Published
|
|
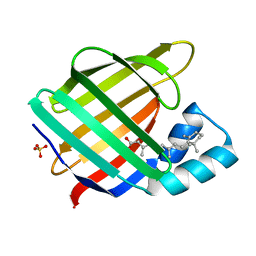
3CWK
 
 | |
3CZE
 
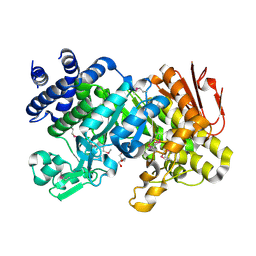 | | Crystal Structure Analysis of Sucrose hydrolase (SUH)- Tris complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Sucrose hydrolase | | Authors: | Kim, M.I, Rhee, S. | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures and mutagenesis of sucrose hydrolase from Xanthomonas axonopodis pv. glycines: insight into the exclusively hydrolytic amylosucrase fold.
J.Mol.Biol., 380, 2008
|
|
3CFY
 
 | | Crystal structure of signal receiver domain of putative Luxo repressor protein from Vibrio parahaemolyticus | | Descriptor: | Putative LuxO repressor protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Fong, R, Freeman, J, Iizuka, M, Groshong, C, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Putative Luxo Repressor Protein from Vibrio Parahaemolyticus.
To be Published
|
|
5KYA
 
 | | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core | | Descriptor: | Oxysterols receptor LXR-beta, Retinoic acid receptor RXR-beta, [2-[(6~{R})-2-(3-methylsulfonylphenyl)-6-propan-2-yl-4,6-dihydropyrrolo[3,4-c]pyrazol-5-yl]-4-(trifluoromethyl)pyrimidin-5-yl]methanol | | Authors: | Chen, G, McKeever, B.M. | | Deposit date: | 2016-07-21 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Brain penetrant liver X receptor (LXR) modulators based on a 2,4,5,6-tetrahydropyrrolo[3,4-c]pyrazole core.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
3CTK
 
 | | Crystal structure of the type 1 RIP bouganin | | Descriptor: | rRNA N-glycosidase | | Authors: | Fermani, S, Tosi, G, Falini, G, Ripamonti, A, Farini, V, Bolognesi, A, Polito, L. | | Deposit date: | 2008-04-14 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure/function studies on two type 1 ribosome inactivating proteins: Bouganin and lychnin.
J.Struct.Biol., 168, 2009
|
|
3D24
 
 | | Crystal structure of ligand-binding domain of estrogen-related receptor alpha (ERRalpha) in complex with the peroxisome proliferators-activated receptor coactivator-1alpha box3 peptide (PGC-1alpha) | | Descriptor: | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Steroid hormone receptor ERR1 | | Authors: | Moras, D, Greschik, H, Flaig, R, Sato, Y, Rochel, N, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-05-07 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Communication between the ERR{alpha} Homodimer Interface and the PGC-1{alpha} Binding Surface via the Helix 8-9 Loop.
J.Biol.Chem., 283, 2008
|
|
3CQO
 
 | |
3CU8
 
 | | Impaired binding of 14-3-3 to Raf1 is linked to Noonan and LEOPARD syndrome | | Descriptor: | 14-3-3 protein zeta/delta, MAGNESIUM ION, PROPANOIC ACID, ... | | Authors: | Schumacher, B, Weyand, M, Kuhlmann, J, Ottmann, C. | | Deposit date: | 2008-04-16 | | Release date: | 2009-05-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling.
Mol. Cell. Biol., 30, 2010
|
|
3CZL
 
 | |
3CSP
 
 | | Crystal structure of the DM2 mutant of myelin oligodendrocyte glycoprotein | | Descriptor: | Myelin-oligodendrocyte glycoprotein | | Authors: | Breithaupt, C, Schafer, B, Pellkofer, H, Huber, R, Linington, C, Jacob, U. | | Deposit date: | 2008-04-10 | | Release date: | 2008-10-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Demyelinating myelin oligodendrocyte glycoprotein-specific autoantibody response is focused on one dominant conformational epitope region in rodents
J.Immunol., 181, 2008
|
|
3CJW
 
 | |
3CZ5
 
 | | Crystal structure of two-component response regulator, LuxR family, from Aurantimonas sp. SI85-9A1 | | Descriptor: | PHOSPHATE ION, Two-component response regulator, LuxR family | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S.R, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-28 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of two-component response regulator, LuxR family, from Aurantimonas sp. SI85-9A1.
To be Published
|
|
8CUR
 
 | | Crystal structure of Cdk2 in complex with Cyclin A inhibitor 6-[(E)-2-(4-chlorophenyl)ethenyl]-2-{[(2R)-3-(4-hydroxyphenyl)-1-methoxy-1-oxopropan-2-yl]carbamoyl}quinoline-4-carboxylic acid | | Descriptor: | 6-[(E)-2-(4-chlorophenyl)ethenyl]-2-{[(2R)-3-(4-hydroxyphenyl)-1-methoxy-1-oxopropan-2-yl]carbamoyl}quinoline-4-carboxylic acid, Cyclin-dependent kinase 2 | | Authors: | Tripathi, S.M, Tambo, C.S, Kiss, G, Rubin, S.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biolayer Interferometry Assay for Cyclin-Dependent Kinase-Cyclin Association Reveals Diverse Effects of Cdk2 Inhibitors on Cyclin Binding Kinetics.
Acs Chem.Biol., 18, 2023
|
|
2N8G
 
 | | NMR Structure of the homeodomain transcription factor Gbx1[E23R,R58E] from Homo sapiens | | Descriptor: | Homeobox protein GBX-1 | | Authors: | Proudfoot, A.K, Serrano, P, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2015-10-15 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the homeodomain transcription factor Gbx1[E23R,R58E] from Homo sapiens
To be Published
|
|
8AFR
 
 | | Pim1 in complex with 4-((6-hydroxybenzofuran-3-yl)methyl)benzoic acid and Pimtide | | Descriptor: | 4-((6-hydroxybenzofuran-3-yl)methyl)benzoic acid, Pimtide, Serine/threonine-protein kinase pim-1 | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2022-07-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
