1VK4
 
 | |
1VM3
 
 | |
1LYB
 
 | | CRYSTAL STRUCTURES OF NATIVE AND INHIBITED FORMS OF HUMAN CATHEPSIN D: IMPLICATIONS FOR LYSOSOMAL TARGETING AND DRUG DESIGN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CATHEPSIN D, PEPSTATIN, ... | | Authors: | Baldwin, E.T, Bhat, T.N, Gulnik, S, Erickson, J.W. | | Deposit date: | 1993-04-22 | | Release date: | 1994-01-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of native and inhibited forms of human cathepsin D: implications for lysosomal targeting and drug design.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1LZZ
 
 | | Rat neuronal NOS heme domain with N-isopropyl-N'-hydroxyguanidine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-ISOPROPYL-N'-HYDROXYGUANIDINE, ... | | Authors: | Li, H, Shimizu, H, Flinspach, M, Jamal, J, Yang, W, Xian, M, Cai, T, Wen, E.Z, Jia, Q, Wang, P.G, Poulos, T.L. | | Deposit date: | 2002-06-11 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Novel Binding Mode of N-Alkyl-N'-Hydroxyguanidine to Neuronal Nitric Oxide
Synthase Provides Mechanistic Insights into NO Biosynthesis
Biochemistry, 41, 2002
|
|
1M0P
 
 | | Structure of Dialkylglycine Decarboxylase Complexed with 1-Amino-1-phenylethanephosphonate | | Descriptor: | (1R)-1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]-1-PHENYLETHYLPHOSPHONIC ACID, 2,2-Dialkylglycine Decarboxylase, POTASSIUM ION, ... | | Authors: | Liu, W, Rogers, C.J, Fisher, A.J, Toney, M.D. | | Deposit date: | 2002-06-13 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aminophosphonate Inhibitors of Dialkylglycine Decarboxylase: Structural Basis for Slow Binding Inhibition
Biochemistry, 41, 2002
|
|
1M23
 
 | |
1LWX
 
 | | AZT DIPHOSPHATE BINDING TO NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Xu, Y. | | Deposit date: | 1997-04-30 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray analysis of azido-thymidine diphosphate binding to nucleoside diphosphate kinase.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1M3D
 
 | | Structure of Type IV Collagen NC1 Domains | | Descriptor: | BROMIDE ION, GLYCEROL, LUTETIUM (III) ION, ... | | Authors: | Sundaramoorthy, M, Meiyappan, M, Todd, P, Hudson, B.G. | | Deposit date: | 2002-06-27 | | Release date: | 2003-01-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of NC1 Domains. Structural Basis for Type IV Collagen Assembly in Basement Membranes
J.Biol.Chem., 277, 2002
|
|
1W0R
 
 | |
1LV4
 
 | | Human catestatin 21-mer | | Descriptor: | catestatin | | Authors: | O'Connor, D.T, Preece, N.E. | | Deposit date: | 2002-05-24 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational preferences and activities of peptides from the catecholamine release-inhibitory (catestatin) region of chromogranin A.
Regul.Pept., 118, 2004
|
|
1SBT
 
 | | ATOMIC COORDINATES FOR SUBTILISIN BPN (OR NOVO) | | Descriptor: | SUBTILISIN BPN' | | Authors: | Alden, R.A, Birktoft, J.J, Kraut, J, Robertus, J.D, Wright, C.S. | | Deposit date: | 1972-08-11 | | Release date: | 1977-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic coordinates for subtilisin BPN' (or Novo).
Biochem.Biophys.Res.Commun., 45, 1971
|
|
1M7W
 
 | | HNF4a ligand binding domain with bound fatty acid | | Descriptor: | Hepatocyte nuclear factor 4-alpha, LAURIC ACID | | Authors: | Dhe-Paganon, S, Duda, K, Iwamoto, M, Chi, Y.I, Shoelson, S.E. | | Deposit date: | 2002-07-22 | | Release date: | 2003-07-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the HNF4 alpha ligand binding domain in complex with endogenous fatty acid ligand
J.Biol.Chem., 277, 2002
|
|
1M0Q
 
 | | Structure of Dialkylglycine Decarboxylase Complexed with S-1-aminoethanephosphonate | | Descriptor: | (1S)-1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]ETHYLPHOSPHONIC ACID, 2,2-Dialkylglycine Decarboxylase, POTASSIUM ION, ... | | Authors: | Liu, W, Rogers, C.J, Fisher, A.J, Toney, M.D. | | Deposit date: | 2002-06-13 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aminophosphonate Inhibitors of Dialkylglycine Decarboxylase: Structural Basis for Slow Binding Inhibition
Biochemistry, 41, 2002
|
|
1M1D
 
 | | TETRAHYMENA GCN5 WITH BOUND BISUBSTRATE ANALOG INHIBITOR | | Descriptor: | HISTONE H3, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Poux, A.N, Cebrat, M, Kim, C.M, Cole, P.A, Marmorstein, R. | | Deposit date: | 2002-06-18 | | Release date: | 2002-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the GCN5 histone acetyltransferase bound to a bisubstrate inhibitor.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1M1Y
 
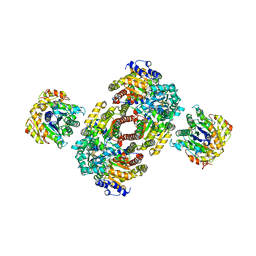 | | Chemical Crosslink of Nitrogenase MoFe Protein and Fe Protein | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Schmid, B, Einsle, O, Chiu, H.J, Willing, A, Yoshida, M, Howard, J.B, Rees, D.C. | | Deposit date: | 2002-06-20 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical and Structural Characterization of the Crosslinked Complex of Nitrogenase: Comparison to the ADP-AlF4- Stabilized Structure
Biochemistry, 41, 2002
|
|
1SYM
 
 | |
1M6V
 
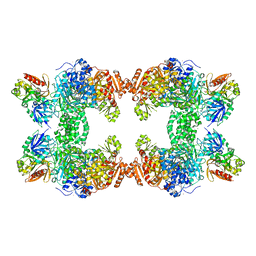 | | Crystal Structure of the G359F (small subunit) Point Mutant of Carbamoyl Phosphate Synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, L-ornithine, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 2002-07-17 | | Release date: | 2002-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carbamoyl-phosphate synthetase. Creation of an escape route for ammonia
J.Biol.Chem., 277, 2002
|
|
1SSC
 
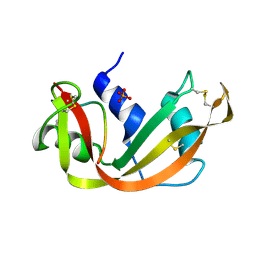 | | THE 1.6 ANGSTROMS STRUCTURE OF A SEMISYNTHETIC RIBONUCLEASE CRYSTALLIZED FROM AQUEOUS ETHANOL. COMPARISON WITH CRYSTALS FROM SALT SOLUTIONS AND WITH RNASE A FROM AQUEOUS ALCOHOL SOLUTIONS | | Descriptor: | PHOSPHATE ION, RIBONUCLEASE A | | Authors: | De Mel, V.S.J, Doscher, M.S, Martin, P.D, Rodier, F, Edwards, B.F.P. | | Deposit date: | 1994-10-05 | | Release date: | 1995-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1.6 A structure of semisynthetic ribonuclease crystallized from aqueous ethanol. Comparison with crystals from salt solutions and with ribonuclease A from aqueous alcohol solutions.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1LZ8
 
 | | LYSOZYME PHASED ON ANOMALOUS SIGNAL OF SULFURS AND CHLORINES | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Dauter, Z, Dauter, M, De La Fortelle, E, Bricogne, G, Sheldrick, G.M. | | Deposit date: | 1999-03-14 | | Release date: | 1999-05-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Can anomalous signal of sulfur become a tool for solving protein crystal structures?
J.Mol.Biol., 289, 1999
|
|
1M0I
 
 | | Crystal Structure of Bacteriophage T7 Endonuclease I with a Wild-Type Active Site | | Descriptor: | SULFATE ION, endodeoxyribonuclease I | | Authors: | Hadden, J.M, Declais, A.C, Phillips, S.E, Lilley, D.M. | | Deposit date: | 2002-06-13 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Metal ions bound at the active site of the junction-resolving enzyme T7 endonuclease I
Embo J., 21, 2002
|
|
1LYA
 
 | | CRYSTAL STRUCTURES OF NATIVE AND INHIBITED FORMS OF HUMAN CATHEPSIN D: IMPLICATIONS FOR LYSOSOMAL TARGETING AND DRUG DESIGN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CATHEPSIN D, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Baldwin, E.T, Bhat, T.N, Gulnik, S, Erickson, J.W. | | Deposit date: | 1993-04-22 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of native and inhibited forms of human cathepsin D: implications for lysosomal targeting and drug design.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1T4N
 
 | | Solution structure of Rnt1p dsRBD | | Descriptor: | Ribonuclease III | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Graille, M, van Tilbeurgh, H, Leeper, T.C, Godin, K.S, Edwards, T.E, Sigurdsson, S.T, Rozenkrants, N, Nagel, R.J, Ares, M, Varani, G. | | Deposit date: | 2004-04-30 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III
Embo J., 23, 2004
|
|
1T4O
 
 | | Crystal structure of rnt1p dsRBD | | Descriptor: | Ribonuclease III | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Graille, M, van Tilbeurgh, H, Leeper, T.C, Godin, K.S, Edwards, T.E, Sigurdsson, S.T, Rozenkrants, N, Nagel, R.J, Ares Jr, M, Varani, G. | | Deposit date: | 2004-04-30 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III
Embo J., 23, 2004
|
|
1T54
 
 | | Antibiotic Activity and Structural Analysis of a Scorpion-derived Antimicrobial peptide IsCT and Its Analogs | | Descriptor: | Cytotoxic linear peptide IsCT | | Authors: | Lee, K, Shin, S.Y, Kim, K, Lim, S.S, Hahm, K.S, Kim, Y. | | Deposit date: | 2004-05-02 | | Release date: | 2004-10-19 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Antibiotic activity and structural analysis of the scorpion-derived antimicrobial peptide IsCT and its analogs
Biochem.Biophys.Res.Commun., 323, 2004
|
|
1M1G
 
 | | Crystal Structure of Aquifex aeolicus N-utilization substance G (NusG), Space Group P2(1) | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Steiner, T, Kaiser, J.T, Marinkovic, S, Huber, R, Wahl, M.C. | | Deposit date: | 2002-06-19 | | Release date: | 2003-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of transcription factor NusG in light of its nucleic
acid- and protein-binding activities
Embo J., 21, 2002
|
|
