7M30
 
 | |
8OPC
 
 | |
7M22
 
 | |
8OPD
 
 | |
7MDO
 
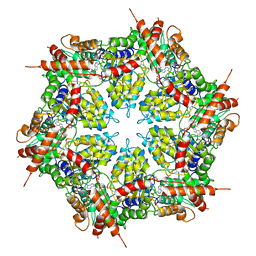 | | Structure of human p97 ATPase L464P mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Zhang, X, Gui, L, Li, S, Nandi, P, Columbres, R.C, Wong, D.E, Moen, D.R, Lin, H.J, Chiu, P.-L, Chou, T.-F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Conserved L464 in p97 D1-D2 linker is critical for p97 cofactor regulated ATPase activity.
Biochem.J., 478, 2021
|
|
7MDM
 
 | | Structure of human p97 ATPase L464P mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Zhang, X, Gui, L, Li, S, Nandi, P, Columbres, R.C, Wong, D.E, Moen, D.R, Lin, H.J, Chiu, P.-L, Chou, T.-F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.86 Å) | | Cite: | Conserved L464 in p97 D1-D2 linker is critical for p97 cofactor regulated ATPase activity.
Biochem.J., 478, 2021
|
|
8OZ0
 
 | | Structure of a human 48S translation initiation complex with eIF4F and eIF4A | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Brito Querido, J, Sokabe, M, Diaz-Lopez, I, Gordiyenko, Y, Fraser, C.S, Ramakrishnan, V. | | Deposit date: | 2023-05-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structure of a human translation initiation complex reveals two independent roles for the helicase eIF4A.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OPB
 
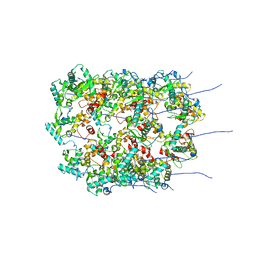 | |
7WBV
 
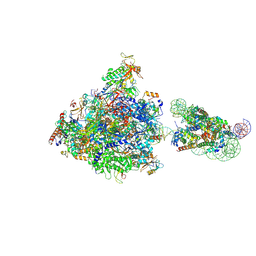 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-4) of the nucleosome | | Descriptor: | DNA (159-MER), DNA (198-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2021-12-17 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
7WBW
 
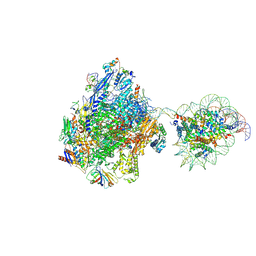 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-3.5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2021-12-17 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
2L15
 
 | | Solution Structure of Cold Shock Protein CspA Using Combined NMR and CS-Rosetta method | | Descriptor: | Cold shock protein CspA | | Authors: | Tang, Y, Schneider, W.M, Shen, Y, Raman, S, Inouye, M, Baker, D, Roth, M.J, Montelione, G.T. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Fully automated high-quality NMR structure determination of small (2)H-enriched proteins.
J Struct Funct Genomics, 11, 2010
|
|
7O2W
 
 | | Structure of the C9orf72-SMCR8 complex | | Descriptor: | Guanine nucleotide exchange protein SMCR8,Guanine nucleotide exchange protein SMCR8,Maltose/maltodextrin-binding periplasmic protein, Ubiquitin-like protein SMT3,Guanine nucleotide exchange C9orf72 | | Authors: | Noerpel, J, Cavadini, S, Schenk, A.D, Graff-Meyer, A, Chao, J, Bhaskar, V. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structure of the human C9orf72-SMCR8 complex reveals a multivalent protein interaction architecture.
Plos Biol., 19, 2021
|
|
5HVC
 
 | | Solution structure of the apo state of the acyl carrier protein from the MLSA2 subunit of the mycolactone polyketide synthase | | Descriptor: | Type I modular polyketide synthase | | Authors: | Vance, S, Tkachenko, O, Thomas, B, Bassuni, M, Hong, H, Nietlispach, D, Broadhurst, R.W. | | Deposit date: | 2016-01-28 | | Release date: | 2016-03-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Sticky swinging arm dynamics: studies of an acyl carrier protein domain from the mycolactone polyketide synthase.
Biochem.J., 473, 2016
|
|
5HV8
 
 | | Solution structure of an octanoyl- loaded acyl carrier protein domain from module MLSA2 of the mycolactone polyketide synthase. | | Descriptor: | S-[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl] octanethioate, Type I modular polyketide synthase | | Authors: | Vance, S, Tkachenko, O, Thomas, B, Bassuni, M, Hong, H, Nietlispach, D, Broadhurst, R.W. | | Deposit date: | 2016-01-28 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Sticky swinging arm dynamics: studies of an acyl carrier protein domain from the mycolactone polyketide synthase.
Biochem.J., 473, 2016
|
|
4U0X
 
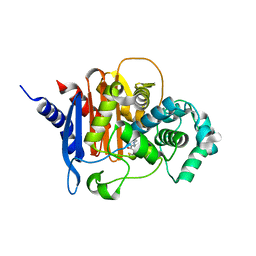 | | Structure of ADC-7 beta-lactamase in complex with boronic acid inhibitor S02030 | | Descriptor: | 1-{(2R)-2-(dihydroxyboranyl)-2-[(thiophen-2-ylacetyl)amino]ethyl}-1H-1,2,3-triazole-4-carboxylic acid, ADC-7 beta-lactamase, PHOSPHATE ION | | Authors: | Powers, R.A, Wallar, B.J, Swanson, H.C. | | Deposit date: | 2014-07-14 | | Release date: | 2014-11-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Biochemical and Structural Analysis of Inhibitors Targeting the ADC-7 Cephalosporinase of Acinetobacter baumannii.
Biochemistry, 53, 2014
|
|
1TUH
 
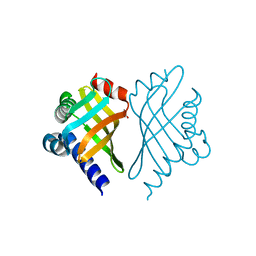 | | Structure of Bal32a from a Soil-Derived Mobile Gene Cassette | | Descriptor: | ACETATE ION, hypothetical protein EGC068 | | Authors: | Robinson, A, Wu, P.S.-C, Harrop, S.J, Schaeffer, P.M, Dixon, N.E, Gillings, M.R, Holmes, A.J, Nevalainen, K.M.H, Otting, G, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2004-06-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Integron-associated Mobile Gene Cassettes Code for Folded Proteins: The Structure of Bal32a, a New Member of the Adaptable alpha+beta Barrel Family
J.Mol.Biol., 346, 2005
|
|
4U0T
 
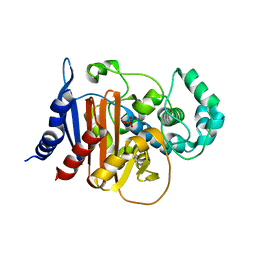 | | Crystal structure of ADC-7 beta-lactamase | | Descriptor: | ADC-7 beta-lactamase, PHOSPHATE ION | | Authors: | Powers, R.A, Wallar, B.J. | | Deposit date: | 2014-07-14 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Biochemical and Structural Analysis of Inhibitors Targeting the ADC-7 Cephalosporinase of Acinetobacter baumannii.
Biochemistry, 53, 2014
|
|
2L6U
 
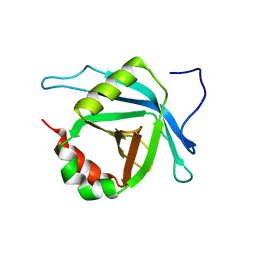 | | Solution NMR Structure of Med25(391-543) Comprising the Activator-Interacting Domain (ACID) of Human Mediator Subuniti 25. Northeast Structural Genomics Consortium Target HR6188A | | Descriptor: | Mediator complex subunit MED25 | | Authors: | Eletsky, A, Ryuechan, W.T, Sukumaran, D.K, Shastry, R, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of MED25(391-543) comprising the activator-interacting domain (ACID) of human mediator subunit 25.
J.Struct.Funct.Genom., 12, 2011
|
|
2LV6
 
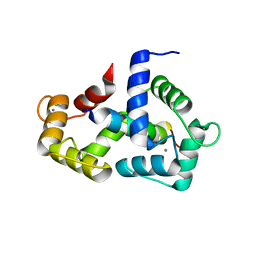 | | The complex between Ca-Calmodulin and skeletal muscle myosin light chain kinase from combination of NMR and aqueous and contrast-matched SAXS data | | Descriptor: | CALCIUM ION, Calmodulin, Myosin light chain kinase 2, ... | | Authors: | Grishaev, A.V, Anthis, N.J, Clore, G.M. | | Deposit date: | 2012-06-29 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Contrast-matched small-angle X-ray scattering from a heavy-atom-labeled protein in structure determination: application to a lead-substituted calmodulin-peptide complex.
J.Am.Chem.Soc., 134, 2012
|
|
2L58
 
 | |
2KAD
 
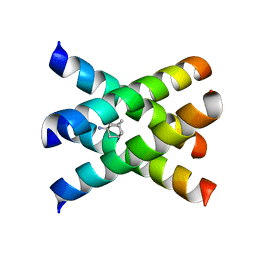 | | Magic-Angle-Spinning Solid-State NMR Structure of Influenza A M2 Transmembrane Domain | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, Transmembrane peptide of Matrix protein 2 | | Authors: | Hong, M, Cady, S.D, Mishanina, T.V. | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure of amantadine-bound M2 transmembrane peptide of influenza A in lipid bilayers from magic-angle-spinning solid-state NMR: the role of Ser31 in amantadine binding.
J.Mol.Biol., 385, 2009
|
|
2LXC
 
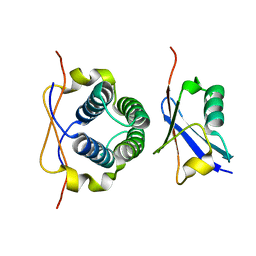 | |
2M0H
 
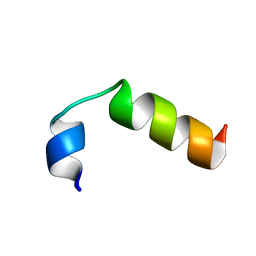 | | SP-B C-terminal (residues 59-80) peptide in methanol | | Descriptor: | Pulmonary surfactant-associated protein B | | Authors: | Kuznetsova, A, Long, J.R. | | Deposit date: | 2012-10-25 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structures of the C-terminal segment of surfactant protein B (residues 59-80) in DPC detergent micelles and methanol.
To be Published
|
|
6PZW
 
 | | CryoEM derived model of NA-22 Fab in complex with N9 Shanghai2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NA-22 fragment antigen binding heavy chain, ... | | Authors: | Ward, A.B, Turner, H.L, Zhu, X. | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Basis of Protection against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies.
Cell Host Microbe, 26, 2019
|
|
6KNG
 
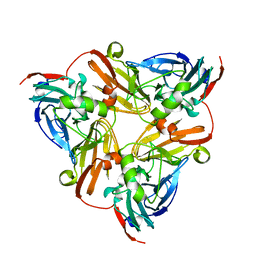 | | CryoEM map and model of Nitrite Reductase at pH 8.1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Adachi, N, Yamaguchi, T, Moriya, T, Kawasaki, M, Koiwai, K, Shinoda, A, Yamada, Y, Yumoto, F, Kohzuma, T, Senda, T. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | 2.85 and 2.99 angstrom resolution structures of 110 kDa nitrite reductase determined by 200 kV cryogenic electron microscopy.
J.Struct.Biol., 213, 2021
|
|
