8B9G
 
 | | Cryo-EM structure of MLE in complex with ADP:AlF4 and U10 RNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dosage compensation regulator, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), ... | | Authors: | Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-10-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis of RNA-induced autoregulation of the DExH-type RNA helicase maleless.
Mol.Cell, 83, 2023
|
|
4PNJ
 
 | | Recombinant Sperm Whale P6 Myoglobin Solved with Single Pulse Free Electron Laser Data | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Cohen, A, Gonzalez, A, Lam, W, Lyubimov, A, Sauter, N, Tsai, Y, Uervirojnangkoorn, M, Brunger, A, Soltis, M. | | Deposit date: | 2014-05-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Goniometer-based femtosecond crystallography with X-ray free electron lasers.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8RCL
 
 | |
8IX0
 
 | | Cryo-EM structure of unprotonated LHCII nanodisc at high pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
8IX2
 
 | | Cryo-EM structure of unprotonated LHCII nanodisc at low pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
8EBU
 
 | | XPC release from Core7-XPA-DNA (Cy5) | | Descriptor: | DNA repair protein complementing XP-A cells, DNA repair protein complementing XP-C cells, DNA1, ... | | Authors: | Kim, J, Yang, W. | | Deposit date: | 2022-08-31 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Lesion recognition by XPC, TFIIH and XPA in DNA excision repair.
Nature, 617, 2023
|
|
8EBY
 
 | | XPC release from Core7-XPA-DNA (AP) | | Descriptor: | DNA, DNA repair protein complementing XP-A cells, DNA repair protein complementing XP-C cells, ... | | Authors: | Kim, J, Yang, W. | | Deposit date: | 2022-08-31 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Lesion recognition by XPC, TFIIH and XPA in DNA excision repair.
Nature, 617, 2023
|
|
8EBV
 
 | |
8IWX
 
 | | Cryo-EM structure of unprotonated LHCII in detergent solution at high pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
8IWY
 
 | | Cryo-EM structure of protonated LHCII in detergent solution at low pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
8EBS
 
 | |
6YUE
 
 | | Fragment of nitrate/nitrite sensor histidine kinase NarQ (R50S variant) | | Descriptor: | Nitrate/nitrite sensor protein NarQ | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Yuzhakova, A, Gordeliy, V. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sensor Histidine Kinase NarQ Activates via Helical Rotation, Diagonal Scissoring, and Eventually Piston-Like Shifts.
Int J Mol Sci, 21, 2020
|
|
8IWZ
 
 | | Cryo-EM structure of unprotonated LHCII in detergent solution at low pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
8EBW
 
 | |
8IX1
 
 | | Cryo-EM structure of protonated LHCII nanodisc at low pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
8R0B
 
 | | Cryo-EM structure of the cross-exon pre-B+ATP complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8R09
 
 | | Cryo-EM structure of the cross-exon pre-B+5'ss+ATPgammaS complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8RM5
 
 | | Cryo-EM structure of the cross-exon pre-B+5'ssLNG+ATPyS complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'SS oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2024-01-05 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8RS9
 
 | | p97 (VCP) double mutant - F266A F539A | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Arie, M, Matzov, D, Karmona, R, Szenkier, N, Stanhill, A, Navon, A. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | p97 (VCP) double mutant - F266A F539A
To Be Published
|
|
7V02
 
 | | Staphylococcus epidermidis RP62A CRISPR short effector complex | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.97 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7UZZ
 
 | | Staphylococcus epidermidis RP62a CRISPR tall effector complex | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7UZW
 
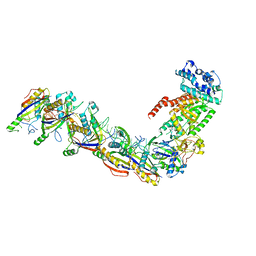 | | Staphylococcus epidermidis RP62a CRISPR effector subcomplex | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm4, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
8IET
 
 | |
7UZX
 
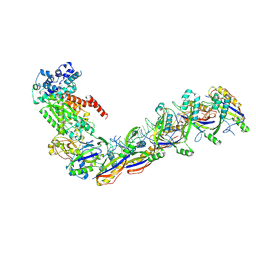 | | Staphylococcus epidermidis RP62a CRISPR effector subcomplex with non-self target RNA bound | | Descriptor: | CRISPR non-self RNA target, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm4, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
8ROZ
 
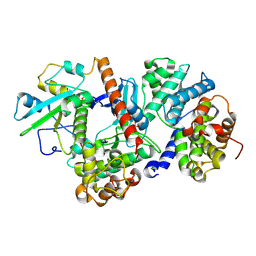 | |
