6GIT
 
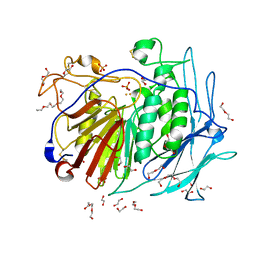 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - PRODUCT COMPLEX | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.418 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6GFV
 
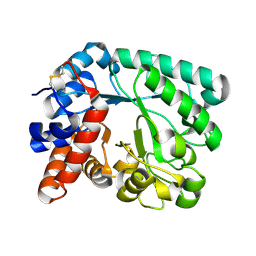 | | M tuberculosis LpqI | | Descriptor: | Probable conserved lipoprotein LpqI | | Authors: | Moynihan, P.J, Lovering, A.L. | | Deposit date: | 2018-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The hydrolase LpqI primes mycobacterial peptidoglycan recycling.
Nat Commun, 10, 2019
|
|
6FFI
 
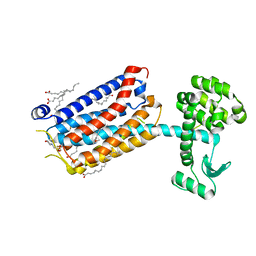 | | Crystal Structure of mGluR5 in complex with MMPEP at 2.2 A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[2-(3-methoxyphenyl)ethynyl]-6-methyl-pyridine, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, ... | | Authors: | Christopher, J.A, Orgovan, Z, Congreve, M, Dore, A.S, Errey, J.C, Marshall, F.H, Mason, J.S, Okrasa, K, Rucktooa, P, Serrano-Vega, M.J, Ferenczy, G.G, Keseru, G.M. | | Deposit date: | 2018-01-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Optimization Strategies for G Protein-Coupled Receptor (GPCR) Allosteric Modulators: A Case Study from Analyses of New Metabotropic Glutamate Receptor 5 (mGlu5) X-ray Structures.
J.Med.Chem., 62, 2019
|
|
6FG6
 
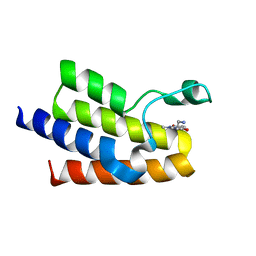 | | Crystal Structure of BAZ2A bromodomain in complex with 1-methylpyridinone compound 1 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, ~{N}-(2-azanylethyl)-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-10 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
6FGH
 
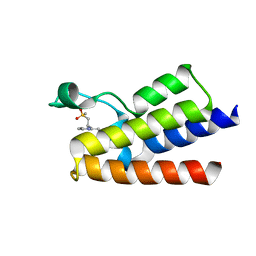 | | Crystal Structure of BAZ2A bromodomain in complex with 3-amino-2-methylpyridine derivative 1 | | Descriptor: | 2-methyl-~{N}-[(2~{R})-1-methylsulfonylpropan-2-yl]pyridin-3-amine, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Marchand, J.-R, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-10 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
6FGL
 
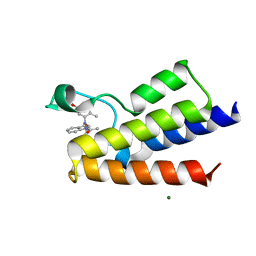 | | Crystal Structure of BAZ2A bromodomain in complex with acetylindole compound UZH47 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, MAGNESIUM ION, N-(1-acetyl-1H-indol-3-yl)-N-(5-hydroxy-2-methylphenyl)acetamide | | Authors: | Dalle Vedove, A, Unzue, A, Nevado, C, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
6FL5
 
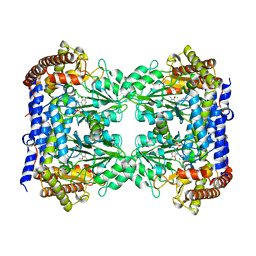 | | Structure of human SHMT1-H135N-R137A-E168N mutant at 3.6 Ang. resolution | | Descriptor: | CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, ... | | Authors: | Giardina, G, Cutruzzola, F, Lucchi, R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The catalytic activity of serine hydroxymethyltransferase is essential for de novo nuclear dTMP synthesis in lung cancer cells.
FEBS J., 285, 2018
|
|
6FGG
 
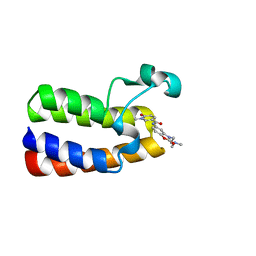 | | Crystal Structure of BAZ2A bromodomain in complex with 1-methylpyridinone compound 5 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, ~{N}-[3-[2-(dimethylamino)ethyl]-2-oxidanylidene-1,3-benzoxazol-5-yl]-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-10 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
6FD9
 
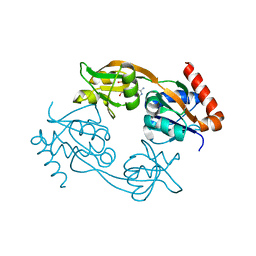 | | Catalytic subunit HisG from Psychrobacter arcticus ATP phosphoribosyltransferase (HisZG ATPPRT) in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP phosphoribosyltransferase | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2017-12-22 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
6FGI
 
 | | Crystal Structure of BAZ2A bromodomain in complex with 3-amino-2-methylpyridine derivative 2 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, ~{N}-[(3~{S})-1,1-bis(oxidanylidene)thian-3-yl]-2-methyl-pyridin-3-amine | | Authors: | Dalle Vedove, A, Marchand, J.-R, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-10 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
6G74
 
 | |
6GD7
 
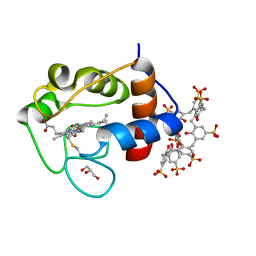 | | Cytochrome c in complex with Sulfonato-calix[8]arene, H3 form with PEG | | Descriptor: | Cytochrome c iso-1, GLYCEROL, HEME C, ... | | Authors: | Rennie, M.L, Fox, G.C, Crowley, P.B. | | Deposit date: | 2018-04-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Auto-regulated Protein Assembly on a Supramolecular Scaffold.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6G8K
 
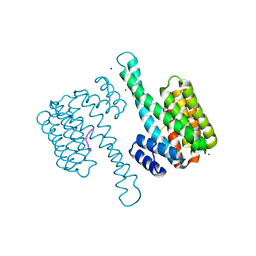 | | 14-3-3sigma in complex with a S131beta3S mutated YAP pS127 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, ACE-ARG-ALA-HIS-SEP-SER-PRO-ALA-BSE-LEU-GLN, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Thijssen, V, Brunsveld, L, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A study on the effect of synthetic alpha-to-beta3-amino acid mutations on the binding of phosphopeptides to 14-3-3 proteins.
Chem.Commun.(Camb.), 55, 2019
|
|
6FSI
 
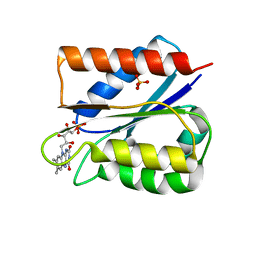 | | Crystal structure of semiquinone Flavodoxin 1 from Bacillus cereus (1.32 A resolution) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, SULFATE ION, ... | | Authors: | Gudim, I, Lofstad, M, Hersleth, H.-P. | | Deposit date: | 2018-02-19 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | High-resolution crystal structures reveal a mixture of conformers of the Gly61-Asp62 peptide bond in an oxidized flavodoxin from Bacillus cereus.
Protein Sci., 27, 2018
|
|
6H3K
 
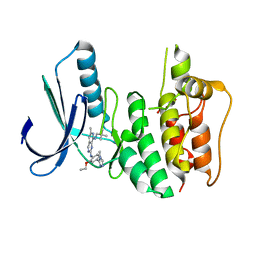 | | Introduction of a methyl group curbs metabolism of pyrido[3,4-d]pyrimidine MPS1 inhibitors and enables the discovery of the Phase 1 clinical candidate BOS172722. | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK, ~{N}8-(2,2-dimethylpropyl)-~{N}2-[2-ethoxy-4-(4-methyl-1,2,4-triazol-3-yl)phenyl]-6-methyl-pyrido[3,4-d]pyrimidine-2,8-diamine | | Authors: | Woodward, H.L, Hoelder, S. | | Deposit date: | 2018-07-19 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Introduction of a Methyl Group Curbs Metabolism of Pyrido[3,4- d]pyrimidine Monopolar Spindle 1 (MPS1) Inhibitors and Enables the Discovery of the Phase 1 Clinical Candidate N2-(2-Ethoxy-4-(4-methyl-4 H-1,2,4-triazol-3-yl)phenyl)-6-methyl- N8-neopentylpyrido[3,4- d]pyrimidine-2,8-diamine (BOS172722).
J. Med. Chem., 61, 2018
|
|
6GHP
 
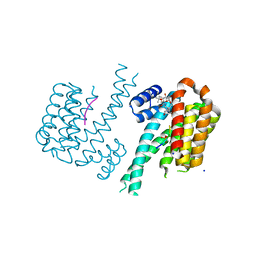 | | 14-3-3sigma in complex with a TASK3 peptide stabilized by semi-synthetic natural product FC-NAc | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Potassium channel subfamily K member 9, ... | | Authors: | Andrei, S.A, de Vink, P.J, Brunsveld, L, Ottmann, C, Higuchi, Y. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-01 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rationally Designed Semisynthetic Natural Product Analogues for Stabilization of 14-3-3 Protein-Protein Interactions.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6FOC
 
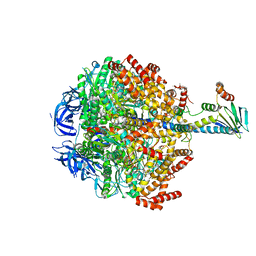 | | F1-ATPase from Mycobacterium smegmatis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | Zhang, T, Montgomery, M.G, Leslie, A.G.W, Cook, G.M, Walker, J.E. | | Deposit date: | 2018-02-06 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The structure of the catalytic domain of the ATP synthase fromMycobacterium smegmatisis a target for developing antitubercular drugs.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6FUC
 
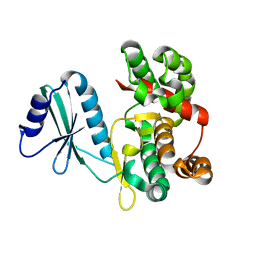 | | Structure of aminoglycoside phosphotransferase APH(3'')-Id from Streptomyces rimosus ATCC10970 | | Descriptor: | Aminoglycoside phosphotransferase | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Alekseeva, M.G, Mavletova, D.A, Zakharevich, N.V, Rudakova, N.N, Danilenko, V.N, Popov, V.O. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Identification, functional and structural characterization of novel aminoglycoside phosphotransferase APH(3′′)-Id from Streptomyces rimosus subsp. rimosus ATCC 10970.
Arch.Biochem.Biophys., 671, 2019
|
|
6GD8
 
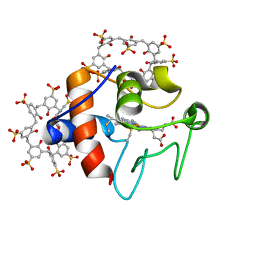 | | Cytochrome c in complex with Sulfonato-calix[8]arene, P31 form | | Descriptor: | Cytochrome c iso-1, HEME C, sulfonato-calix[8]arene | | Authors: | Rennie, M.L, Fox, G.C, Crowley, P.B. | | Deposit date: | 2018-04-23 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Auto-regulated Protein Assembly on a Supramolecular Scaffold.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6G8P
 
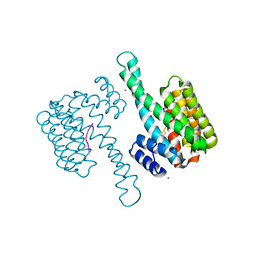 | | 14-3-3sigma in complex with a P129beta3P and L132beta3L mutated YAP pS127 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Thijssen, V, Brunsveld, L, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A study on the effect of synthetic alpha-to-beta3-amino acid mutations on the binding of phosphopeptides to 14-3-3 proteins.
Chem.Commun.(Camb.), 55, 2019
|
|
5TUH
 
 | |
6GBN
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Cytophaga hutchinsonii in complex with adenosine | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Czyrko, J, Jaskolski, M, Brzezinski, K. | | Deposit date: | 2018-04-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from Cytophaga hutchinsonii, a case of combination of crystallographic and non-crystallographic symmetry.
Croatica Chemica Acta, 91, 2018
|
|
6FU2
 
 | | ATP phosphoribosyltransferase (HisZG ATPPRT) from Psychrobacter arcticus in complex with PRPP and ATP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2018-02-26 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
6FU7
 
 | | ATP phosphoribosyltransferase (HisZG ATPPRT) from Psychrobacter arcticus in complex with PRATP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP phosphoribosyltransferase, ATP phosphoribosyltransferase regulatory subunit, ... | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2018-02-26 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
6GL0
 
 | | Structure of ZgEngAGH5_4 in complex with a cellotriose | | Descriptor: | Endoglucanase, family GH5, MAGNESIUM ION, ... | | Authors: | Dorival, J, Ruppert, S, Gunnoo, M, Orlowski, A, Chapelais, M, Dabin, J, Labourel, A, Thompson, D, Michel, G, Czjzek, M, Genicot, S. | | Deposit date: | 2018-05-22 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The laterally acquired GH5ZgEngAGH5_4from the marine bacteriumZobellia galactanivoransis dedicated to hemicellulose hydrolysis.
Biochem. J., 475, 2018
|
|
