3RMA
 
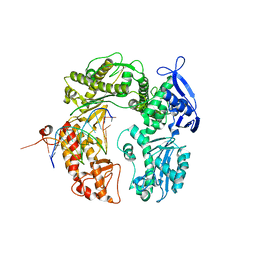 | | Crystal Structure of a replicative DNA polymerase bound to DNA containing Thymine Glycol | | Descriptor: | DNA (5'-D(*CP*GP*AP*(CTG)*GP*AP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*TP*CP*A)-3'), DNA polymerase | | Authors: | Aller, P, Duclos, S, Wallace, S.S, Doublie, S. | | Deposit date: | 2011-04-20 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | A crystallographic study of the role of sequence context in thymine glycol bypass by a replicative DNA polymerase serendipitously sheds light on the exonuclease complex.
J.Mol.Biol., 412, 2011
|
|
3RNY
 
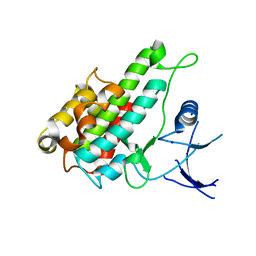 | | Crystal structure of human RSK1 C-terminal kinase domain | | Descriptor: | Ribosomal protein S6 kinase alpha-1, SODIUM ION | | Authors: | Li, D, Fu, T.-M, Nan, J, Su, X.-D. | | Deposit date: | 2011-04-24 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the autoinhibition of the C-terminal kinase domain of human RSK1.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3RQZ
 
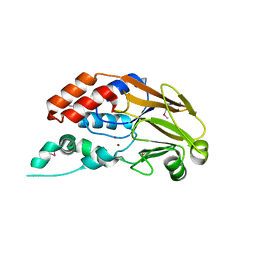 | | Crystal structure of metallophosphoesterase from Sphaerobacter thermophilus | | Descriptor: | ACETATE ION, Metallophosphoesterase, ZINC ION | | Authors: | Chang, C, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-28 | | Release date: | 2011-05-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of metallophosphoesterase from Sphaerobacter thermophilus
To be Published
|
|
3R3R
 
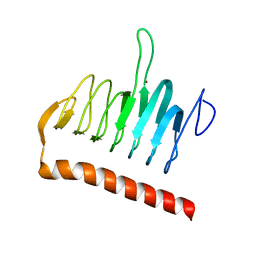 | | Structure of the YrdA ferripyochelin binding protein from Salmonella enterica | | Descriptor: | ZINC ION, ferripyochelin binding protein | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-16 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the YrdA ferripyochelin binding protein from Salmonella enterica
TO BE PUBLISHED
|
|
6TMR
 
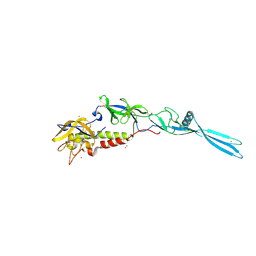 | | Mokola virus glycoprotein, monomeric post-fusion conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, TERBIUM(III) ION | | Authors: | Belot, L, Roche, S, Legrand, P, Gaudin, Y, Albertini, A. | | Deposit date: | 2019-12-05 | | Release date: | 2020-02-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Crystal structure of Mokola virus glycoprotein in its post-fusion conformation.
Plos Pathog., 16, 2020
|
|
8DS5
 
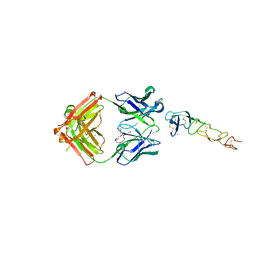 | | X-ray structure of the MK5890 Fab - CD27 antibody-antigen complex | | Descriptor: | CADMIUM ION, CD27 antigen, MK-5890 Fab heavy chain, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Preclinical characterization and clinical translation of pharmacodynamic markers for MK-5890: a human CD27 activating antibody for cancer immunotherapy.
J Immunother Cancer, 10, 2022
|
|
6TPG
 
 | | Crystal structure of the Orexin-2 receptor in complex with EMPA at 2.74 A resolution | | Descriptor: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Hypocretin receptor-2, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.741 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6T7U
 
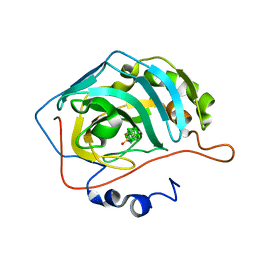 | | Carborane inhibitor of Carbonic Anhydrase IX | | Descriptor: | Carbonic anhydrase 2, Carborane inhibitor, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Kugler, M, Gruner, B. | | Deposit date: | 2019-10-23 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Sulfonamido carboranes as highly selective inhibitors of cancer-specific carbonic anhydrase IX.
Eur.J.Med.Chem., 200, 2020
|
|
3RQ6
 
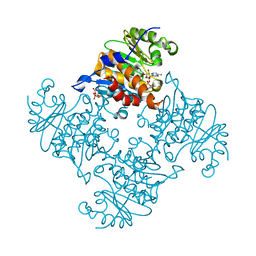 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis soaked with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP/ATP-dependent NAD(P)H-hydrate dehydratase, MAGNESIUM ION | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RFA
 
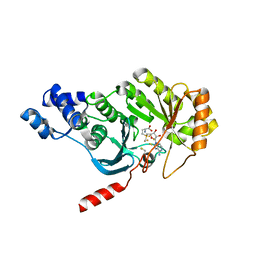 | | X-ray structure of RlmN from Escherichia coli in complex with S-adenosylmethionine | | Descriptor: | IRON/SULFUR CLUSTER, Ribosomal RNA large subunit methyltransferase N, S-ADENOSYLMETHIONINE | | Authors: | Boal, A.K, Grove, T.L, McLaughlin, M.I, Yennawar, N, Booker, S.J, Rosenzweig, A.C. | | Deposit date: | 2011-04-05 | | Release date: | 2011-05-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for methyl transfer by a radical SAM enzyme.
Science, 332, 2011
|
|
3T32
 
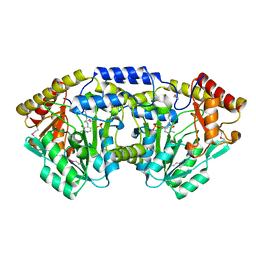 | | Crystal structure of a putative C-S lyase from Bacillus anthracis | | Descriptor: | Aminotransferase, class I/II | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Peterson, S.N, Porebski, P, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-24 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative C-S lyase from Bacillus anthracis
TO BE PUBLISHED
|
|
6T3C
 
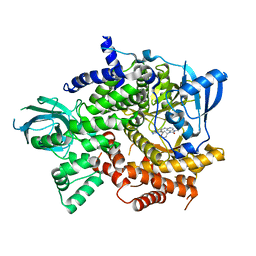 | | Crystal structure of PI3Kgamma in complex with DNA-PK inhibitor AZD7648 | | Descriptor: | 7-methyl-2-[(7-methyl-[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(oxan-4-yl)purin-8-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Schimpl, M, Goldberg, F.W, Finlay, M.R.V, Ting, A.K.T, Beattie, D, Lamont, G.M, Fallan, C, Wrigley, G.L, Howard, M.R, Williamson, B, Davies, B.R, Cadogan, E.B, Ramos-Montoya, A, Dean, E. | | Deposit date: | 2019-10-10 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The Discovery of 7-Methyl-2-[(7-methyl[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8-one (AZD7648), a Potent and Selective DNA-Dependent Protein Kinase (DNA-PK) Inhibitor.
J.Med.Chem., 63, 2020
|
|
6T3T
 
 | | Structure of the 4-hydroxy-tetrahydrodipicolinate synthase from the thermoacidophilic methanotroph Methylacidiphilum fumariolicum SolV | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, SULFATE ION | | Authors: | Schmitz, R, Dietl, A, Mueller, M, Berben, T, Op den Camp, H, Barends, T. | | Deposit date: | 2019-10-11 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the 4-hydroxy-tetrahydrodipicolinate synthase from the thermoacidophilic methanotroph Methylacidiphilum fumariolicum SolV and the phylogeny of the aminotransferase pathway.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
3T65
 
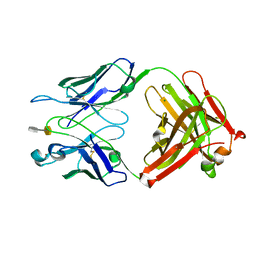 | | S25-2- A(2-8)KDO disaccharide complex | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, MAGNESIUM ION, S25-2 FAB (IGG1K) heavy chain, ... | | Authors: | Nguyen, H.P, Seto, N.O, Mackenzie, C.R, Brade, L, Kosma, P, Brade, H, Evans, S.V. | | Deposit date: | 2011-07-28 | | Release date: | 2011-08-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Germline antibody recognition of distinct carbohydrate epitopes.
Nat.Struct.Biol., 10, 2003
|
|
3SPE
 
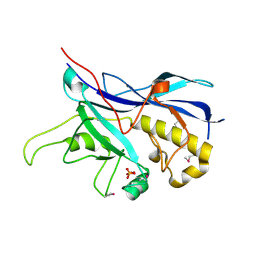 | | Crystal structure of the tail sheath protein protease resistant fragment from bacteriophage phiKZ | | Descriptor: | GLYCEROL, PHIKZ029, PHOSPHATE ION | | Authors: | Aksyuk, A.A, Kurochkina, L.P, Fokine, A, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2011-07-01 | | Release date: | 2011-12-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3996 Å) | | Cite: | Structural conservation of the myoviridae phage tail sheath protein fold.
Structure, 19, 2011
|
|
7QOX
 
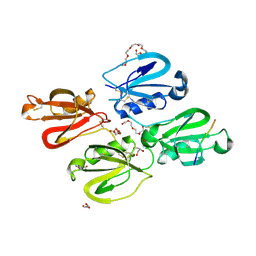 | | Factor XI and Plasma Kallikrein apple domain structures reveals different kininogen bound complexes | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Li, C, Awital, B, Wong, S, Dreveny, I, Meijers, J, Emsley, J. | | Deposit date: | 2021-12-29 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Plasma kallikrein structure reveals apple domain disc rotated conformation compared to factor XI.
J Thromb Haemost, 17, 2019
|
|
7QOT
 
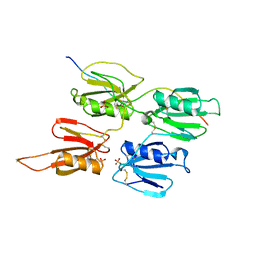 | | Factor XI and Plasma Kallikrein apple domain structures reveals different kininogen bound complexes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa heavy chain, Kininogen-1 light chain, ... | | Authors: | Li, C, Awital, B, Wong, S, Dreveny, I, Meijers, J, Emsley, J. | | Deposit date: | 2021-12-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Plasma kallikrein structure reveals apple domain disc rotated conformation compared to factor XI.
J Thromb Haemost, 17, 2019
|
|
3STF
 
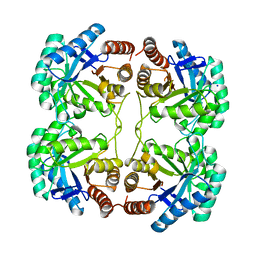 | | Crystal structure of a mutant (S211A) of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Allison, T.M, Jameson, G.B, Gloyne, B.J, Parker, E.J. | | Deposit date: | 2011-07-09 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An Extended (beta)7(alpha)7 Substrate-Binding Loop Is Essential for Efficient Catalysis by 3-Deoxy-D-manno-Octulosonate 8-Phosphate Synthase
Biochemistry, 50, 2011
|
|
7QTZ
 
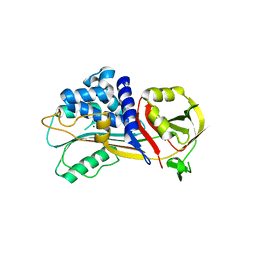 | | Crystal structure of Iripin-1 serpin from tick Ixodes ricinus | | Descriptor: | MAGNESIUM ION, Putative salivary serpin | | Authors: | Kascakova, B, Kuta Smatanova, I, Chmelar, J, Prudnikova, T. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iripin-1, a new anti-inflammatory tick serpin, inhibits leukocyte recruitment in vivo while altering the levels of chemokines and adhesion molecules.
Front Immunol, 14, 2023
|
|
7QXV
 
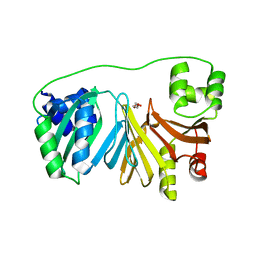 | | Crystal Structure of Haem-Binding Protein HemS Mutant F104A F199A, from Yersinia enterocolitica | | Descriptor: | DI(HYDROXYETHYL)ETHER, Hemin transport protein | | Authors: | Barker, P.D, Keith, A, Brear, P, Wales, D. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal Structure of Haem-Binding Protein HemS Mutant F104A F199A, from Yersinia enterocolitica
To Be Published
|
|
6SRE
 
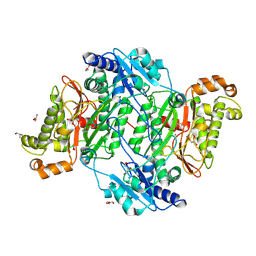 | | Crystal Structure of Human Prolidase S202F variant expressed in the presence of chaperones | | Descriptor: | GLYCEROL, GLYCINE, MANGANESE (II) ION, ... | | Authors: | Wator, E, Rutkiewicz, M, Wilk, P. | | Deposit date: | 2019-09-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Co-expression with chaperones can affect protein 3D structure as exemplified by loss-of-function variants of human prolidase.
Febs Lett., 594, 2020
|
|
3SS7
 
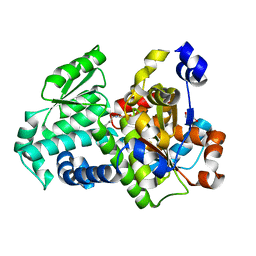 | | Crystal structure of holo D-serine dehydratase from Escherichia coli at 1.55 A resolution | | Descriptor: | D-serine dehydratase, GLYCEROL, POTASSIUM ION, ... | | Authors: | Urusova, D.V, Isupov, M.N, Antonyuk, S.V, Kachalova, G.S, Vagin, A.A, Lebedev, A.A, Bourenkov, G.P, Dauter, Z, Bartunik, H.D, Melik-Adamyan, W.R, Mueller, T.D, Schnackerz, K.D. | | Deposit date: | 2011-07-07 | | Release date: | 2012-01-18 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of D-serine dehydratase from Escherichia coli.
Biochim.Biophys.Acta, 1824, 2011
|
|
6SKC
 
 | | Crystal Structure of Human Kallikrein 6 (I218Y) in complex with GSK3448330A | | Descriptor: | 4-[(3~{S})-1-oxidanyl-3,4-dihydro-2,1-benzoxaborinin-3-yl]benzenecarboximidamide, BENZAMIDINE, GLYCEROL, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-08-15 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Design and development of a series of borocycles as selective, covalent kallikrein 5 inhibitors.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6SX0
 
 | | Specific dsRNA recognition by wild type H7N1 NS1 RNA-binding domain | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Non-structural protein 1, ... | | Authors: | Coste, F, Wacquiez, A, Marc, D, Castaing, B. | | Deposit date: | 2019-09-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Sequence Determinants Governing the Interactions of RNAs with Influenza A Virus Non-Structural Protein NS1.
Viruses, 12, 2020
|
|
6SYH
 
 | |
