7KGT
 
 | | Crystal Structure of HLA-A*0201 in complex with SARS-CoV-2 N226-234 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A*02:01 molecule.
Iscience, 24, 2021
|
|
7C53
 
 | |
7KZB
 
 | |
6F9Y
 
 | |
7KGO
 
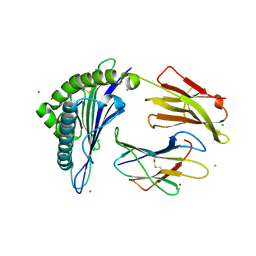 | | Crystal Structure of HLA-A*0201in complex with SARS-CoV-2 N351-359 | | Descriptor: | Beta-2-microglobulin, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Szeto, C, Chatzileontiadou, D.S.M, Riboldi-Tunnicliffe, A, Gras, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The presentation of SARS-CoV-2 peptides by the common HLA-A * 02:01 molecule.
Iscience, 24, 2021
|
|
7KZC
 
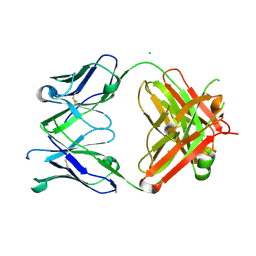 | |
7KZA
 
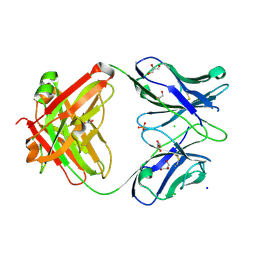 | |
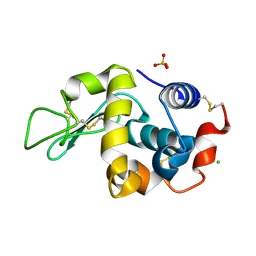
6F1L
 
 | |
6F1H
 
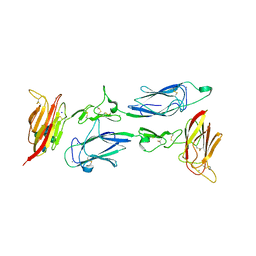 | | C1rC1s complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-galactopyranose-(1-4)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Almitairi, J.O.M, Venkatraman Girija, U, Furze, C.M, Simpson-Gray, X, Badakshi, F, Marshall, J.E, Mitchell, D.A, Moody, P.C.E, Wallis, R. | | Deposit date: | 2017-11-22 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of the C1r-C1s interaction of the C1 complex of complement activation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7CWK
 
 | |
6F1P
 
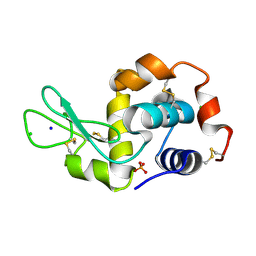 | |
6F7X
 
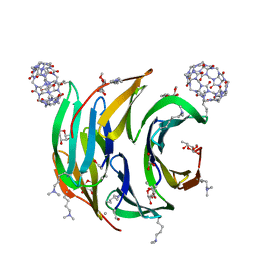 | | Crystal structure of dimethylated RSL - cucurbit[7]uril complex, F432 form | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, cucurbit[7]uril, ... | | Authors: | Guagnini, F, Rennie, M.L, Crowley, P.B. | | Deposit date: | 2017-12-12 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Cucurbit[7]uril-Dimethyllysine Recognition in a Model Protein.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7L4L
 
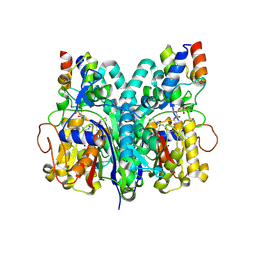 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabF, and C8-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, Acyl carrier protein, SODIUM ION, ... | | Authors: | Mindrebo, J.T, Chen, A, Kim, W.E, Burkart, M.D, Noel, J.P. | | Deposit date: | 2020-12-19 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and Mechanistic Analyses of the Gating Mechanism of Elongating Ketosynthases
Acs Catalysis, 11, 2021
|
|
6F4L
 
 | |
7L4E
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabF, and C16:1-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier-protein] synthase 2, Acyl carrier protein, ... | | Authors: | Mindrebo, J.T, Chen, A, Kim, W.E, Burkart, M.D, Noel, J.P. | | Deposit date: | 2020-12-18 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Mechanistic Analyses of the Gating Mechanism of Elongating Ketosynthases
Acs Catalysis, 11, 2021
|
|
6F9Z
 
 | |
6FAU
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2e-I with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{R})-(2-methoxyphenyl)-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ACE-ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6FI4
 
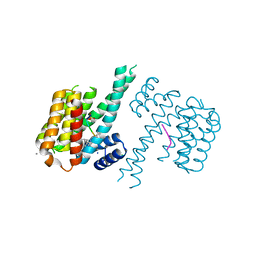 | | Crystal structure of C-terminal modified Tau peptide-hybrid 3.2e with 14-3-3sigma | | Descriptor: | (2~{S})-2-(diphenylmethyl)pyrrolidine, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-01-17 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6FCA
 
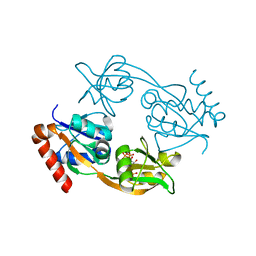 | | Catalytic subunit HisG from Psychrobacter arcticus ATP phosphoribosyltransferase (HisZG ATPPRT) in complex with PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ATP phosphoribosyltransferase | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2017-12-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
7KZG
 
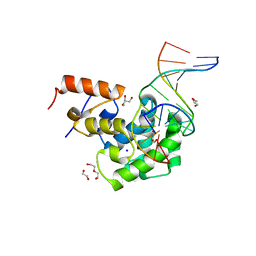 | | Human MBD4 glycosylase domain bound to DNA containing oxacarbenium-ion analog 1-aza-2'-deoxyribose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | Deposit date: | 2020-12-10 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Insights into the Mechanism of Base Excision by MBD4.
J.Mol.Biol., 433, 2021
|
|
7KZ1
 
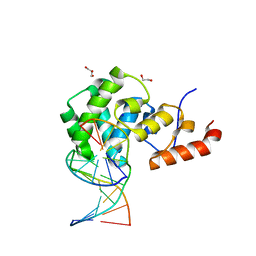 | | Human MBD4 glycosylase domain bound to DNA containing an abasic site | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*CP*GP*(ORP)P*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*GP*CP*GP*CP*TP*GP*G)-3'), ... | | Authors: | Pidugu, L.S, Bright, H, Pozharski, E, Drohat, A.C. | | Deposit date: | 2020-12-09 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Insights into the Mechanism of Base Excision by MBD4.
J.Mol.Biol., 433, 2021
|
|
6F1M
 
 | |
6FAV
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2f-I with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{R})-(3-methoxyphenyl)-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ACE-ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
7KQK
 
 | |
6FCC
 
 | | Catalytic subunit HisG from Psychrobacter arcticus ATP phosphoribosyltransferase (HisZG ATPPRT) | | Descriptor: | ATP phosphoribosyltransferase, L(+)-TARTARIC ACID | | Authors: | Alphey, M.S, Ge, Y, Fisher, G, Czekster, C.M, Naismith, J.H, da Silva, R.G. | | Deposit date: | 2017-12-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Catalytic and Anticatalytic Snapshots of a Short-Form ATP Phosphoribosyltransferase
Acs Catalysis, 2018
|
|
