5WEE
 
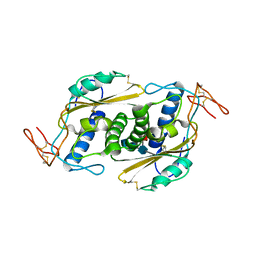 | | Crystal structure of HpVAL4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Asojo, O.A. | | Deposit date: | 2017-07-09 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Heligmosomoides polygyrus Venom Allergen-like Protein-4 (HpVAL-4) is a sterol binding protein.
Int. J. Parasitol., 48, 2018
|
|
5WEP
 
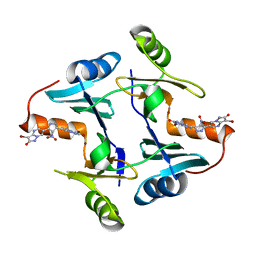 | | Crystal structure of fosfomycin resistance protein FosA3 with inhibitor (ANY1) bound | | Descriptor: | 6,6'-(4-nitro-1H-pyrazole-3,5-diyl)bis(3-bromopyrazolo[1,5-a]pyrimidin-2(1H)-one), FosA3, ZINC ION | | Authors: | Klontz, E.H, Sundberg, E.J. | | Deposit date: | 2017-07-10 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Small-Molecule Inhibitor of FosA Expands Fosfomycin Activity to Multidrug-Resistant Gram-Negative Pathogens.
Antimicrob. Agents Chemother., 63, 2019
|
|
5EV8
 
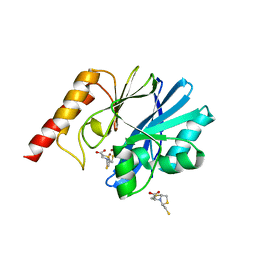 | | Crystal structure of the metallo-beta-lactamase IMP-1 in complex with the bisthiazolidine inhibitor D-CS319 | | Descriptor: | (3S,5S,7aR)-5-(sulfanylmethyl)tetrahydro[1,3]thiazolo[4,3-b][1,3]thiazole-3-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase IMP-1, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cross-class metallo-beta-lactamase inhibition by bisthiazolidines reveals multiple binding modes.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5H2X
 
 | |
5WGR
 
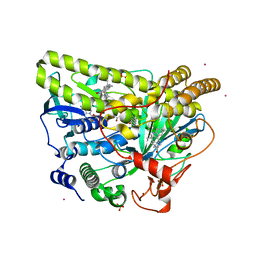 | | Crystal Structure of Wild-type MalA', premalbrancheamide complex | | Descriptor: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Fraley, A.E, Smith, J.L. | | Deposit date: | 2017-07-14 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.362 Å) | | Cite: | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5WGZ
 
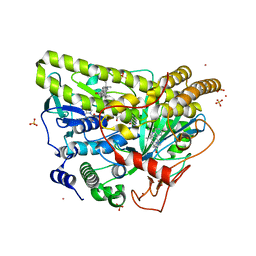 | | Crystal Structure of Wild-type MalA', isomalbrancheamide B complex | | Descriptor: | (5aS,12aS,13aS)-8-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Fraley, A.E, Smith, J.L. | | Deposit date: | 2017-07-14 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5G4Z
 
 | | Structural basis for carboxylic acid recognition by a Cache chemosensory domain. | | Descriptor: | Methyl-accepting chemotaxis sensory transducer with Cache sensor, TRIETHYLENE GLYCOL, UNKNOWN LIGAND | | Authors: | Brewster, J, McKellar, J.L.O, Newman, J, Peat, T.S, Gerth, M.L. | | Deposit date: | 2016-05-18 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for ligand recognition by a Cache chemosensory domain that mediates carboxylate sensing in Pseudomonas syringae.
Sci Rep, 6, 2016
|
|
5G5V
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-038 | | Descriptor: | (4AS,8AR)-4-(3-{4-[(3R)-3-HYDROXYPYRROLIDINE-1-, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2016-06-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
7EQL
 
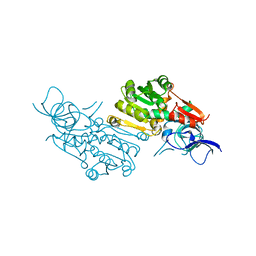 | | Crystal structure of (+)-pulegone reductase from Mentha piperita | | Descriptor: | (+)-pulegone reductase | | Authors: | Lin, W. | | Deposit date: | 2021-05-03 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Functional Characterization and Structural Insights Into Stereoselectivity of Pulegone Reductase in Menthol Biosynthesis.
Front Plant Sci, 12, 2021
|
|
5EZ9
 
 | |
5EZH
 
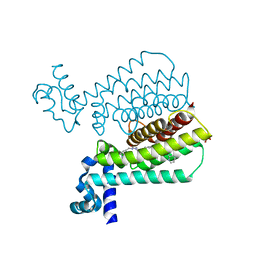 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 21 at 1.7A resolution | | Descriptor: | HTH-type transcriptional regulator EthR, SULFATE ION, ~{N}-[(1-pyridin-2-ylpiperidin-4-yl)methyl]pyrrolidine-1-carboxamide | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-26 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
5WJ4
 
 | |
7ESW
 
 | |
5WJK
 
 | | 2.0-Angstrom In situ Mylar structure of sperm whale myoglobin (SWMb) at 293 K | | Descriptor: | CHLORIDE ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Broecker, J, Ou, W.-L, Ernst, O.P. | | Deposit date: | 2017-07-23 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-throughput in situ X-ray screening of and data collection from protein crystals at room temperature and under cryogenic conditions.
Nat Protoc, 13, 2018
|
|
5GIV
 
 | | Crystal structure of M32 carboxypeptidase from Deinococcus radiodurans R1 | | Descriptor: | ACETATE ION, Carboxypeptidase 1, ZINC ION | | Authors: | Sharma, B, Singh, R, Yadav, P, Ghosh, B, Kumar, A, Jamdar, S.N, Makde, R.D. | | Deposit date: | 2016-06-25 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Active site gate of M32 carboxypeptidases illuminated by crystal structure and molecular dynamics simulations
Biochim. Biophys. Acta, 1865, 2017
|
|
5EZB
 
 | | Chicken prestin STAS domain | | Descriptor: | Chicken prestin STAS domain,Chicken prestin STAS domain, GLYCEROL, OXALATE ION, ... | | Authors: | Lolli, G, Pasqualetto, E, Costanzi, E, Bonetto, G, Battistutta, R. | | Deposit date: | 2015-11-26 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The STAS domain of mammalian SLC26A5 prestin harbours an anion-binding site.
Biochem.J., 473, 2016
|
|
5WGI
 
 | |
5WGX
 
 | | Crystal Structure of MalA' H253A, malbrancheamide B complex | | Descriptor: | (5aS,12aS,13aS)-9-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Fraley, A.E, Smith, J.L. | | Deposit date: | 2017-07-14 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5F25
 
 | | Crystal structure of the BRD9 bromodomain in complex with compound 4. | | Descriptor: | 4-(1,5-dimethyl-6-oxidanylidene-pyridin-3-yl)benzamide, BRD9 | | Authors: | Bader, G, Martin, L.J, Steurer, S, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2015-12-01 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
5F0F
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with compound 15 at 1.76A resolution | | Descriptor: | 4-[4-(3-oxidanylidene-3-pyrrolidin-1-yl-propyl)piperidin-1-yl]benzenecarbonitrile, HTH-type transcriptional regulator EthR, SULFATE ION | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
5WMN
 
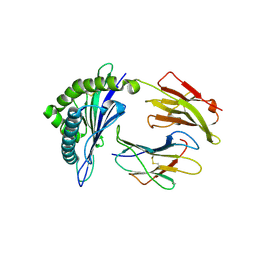 | | Crystal Structure of HLA-B7 in complex with SPI, an influenza peptide | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-30 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Inability To Detect Cross-Reactive Memory T Cells Challenges the Frequency of Heterologous Immunity among Common Viruses.
J. Immunol., 200, 2018
|
|
5IED
 
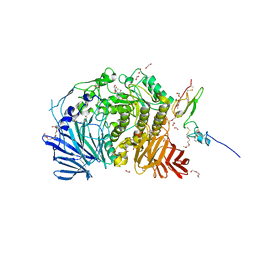 | | Murine endoplasmic reticulum alpha-glucosidase II with castanospermine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-02-25 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F2S
 
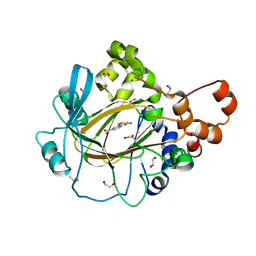 | | Crystal structure of human KDM4A in complex with compound 15 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-azanyl-1,3-thiazol-4-yl)pyridine-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Le Bihan, Y.-V, Dempster, S, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5WLY
 
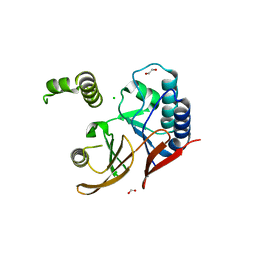 | | E. coli LpxH- 8 mutations | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | Deposit date: | 2017-07-28 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The substrate-binding cap of the UDP-diacylglucosamine pyrophosphatase LpxH is highly flexible, enabling facile substrate binding and product release.
J. Biol. Chem., 293, 2018
|
|
5F3C
 
 | | Crystal structure of human KDM4A in complex with compound 52d | | Descriptor: | 8-[4-[2-[(4-fluorophenyl)methyl-methyl-amino]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.-V, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
