5FBE
 
 | | COMPLEMENT FACTOR D IN COMPLEX WITH COMPOUND2 | | Descriptor: | Complement factor D, GLYCEROL, methyl 2-[[[(2~{S})-2-[[3-(trifluoromethyloxy)phenyl]carbamoyl]pyrrolidin-1-yl]carbonylamino]methyl]benzoate | | Authors: | Ostermann, N, Zink, F. | | Deposit date: | 2015-12-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Small-molecule factor D inhibitors targeting the alternative complement pathway.
Nat.Chem.Biol., 12, 2016
|
|
7AXV
 
 | | Crystal structure of the cAMP-dependent protein kinase A cocrystallized with 5-isoquinolinesulfonic acid and PKI (5-24) | | Descriptor: | DIMETHYL SULFOXIDE, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha, ... | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2020-11-10 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment based drug design - Small chemical changes of fragments effecting big changes in binding
To Be Published
|
|
7AXT
 
 | |
7AY9
 
 | | Crystal structure of CK2 bound by compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 7-(cyclopropylamino)-5-(5-(6-oxo-1,6-dihydropyridin-3-yl)-1-(2-(piperidin-1-yl)ethyl)-1H-1,2,3-triazol-4-yl)pyrazolo[1,5-a]pyrimidine-3-carbonitrile, Casein kinase II subunit alpha, ... | | Authors: | Ferguson, A, Collie, G.W. | | Deposit date: | 2020-11-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Metadynamics simulations of CK2 compound unbinding to understand slow dissociation kinetics.
To Be Published
|
|
4ROM
 
 | | Deoxyhemoglobin in complex with imidazolylacryloyl derivatives | | Descriptor: | 4-{2-chloro-4-[3-(1H-imidazol-2-yl)propanoyl]phenoxy}butanoic acid, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Omar, A.M, Mahran, M.A, Ghatge, M.N, Chowdhury, N, Bamane, F.H.A, El-Araby, M.E, Abdulmalik, O, Safo, M.K. | | Deposit date: | 2014-10-28 | | Release date: | 2015-05-20 | | Last modified: | 2015-06-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a novel class of covalent modifiers of hemoglobin as potential antisickling agents.
Org.Biomol.Chem., 13, 2015
|
|
7JWX
 
 | | Crystal Structure of Trypsin Bound O-methyl Benzamidine | | Descriptor: | 4-[(1-{(1S,2S)-1-[1-(4-aminobutyl)-1H-1,2,3-triazol-4-yl]-2-methylbutyl}-1H-1,2,3-triazol-4-yl)methoxy]-3-methoxybenzene-1-carboximidamide, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Packianathan, C, Laganowsky, A. | | Deposit date: | 2020-08-26 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Small molecule peptidomimetic trypsin inhibitors: validation of an EKO binding mode, but with a twist.
Org.Biomol.Chem., 20, 2022
|
|
7AYA
 
 | | Crystal structure of CK2 bound by compound 9 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Ferguson, A, Collie, G.W. | | Deposit date: | 2020-11-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Metadynamics simulations of CK2 compound unbinding to understand slow dissociation kinetics.
To Be Published
|
|
8CHH
 
 | |
8CCO
 
 | |
7AXW
 
 | |
8CIK
 
 | | Altai wapiti (Cervus elaphus sibiricus) chymosin at 2.2 A resolution | | Descriptor: | Chymosin, GLYCEROL | | Authors: | Diusenova, S.E, Shevtsov, M.B, Borshchevskiy, V.I, Belenkaya, S.V, Kolybalov, D.S, Arkhipov, S.G, Volosnikova, E.A, Elchaninov, V.V, Shcherbakov, D.N. | | Deposit date: | 2023-02-09 | | Release date: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Altai wapiti (Cervus elaphus sibiricus) chymosin at 2.2 A resolution
To Be Published
|
|
4RKE
 
 | | Drosophila melanogaster Rab2 bound to GMPPNP | | Descriptor: | GH01619p, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Lardong, J.A, Driller, J.H, Depner, H, Weise, C, Petzoldt, A, Wahl, M.C, Sigrist, S.J, Loll, B. | | Deposit date: | 2014-10-13 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.0006 Å) | | Cite: | Structures of Drosophila melanogaster Rab2 and Rab3 bound to GMPPNP.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
4RRO
 
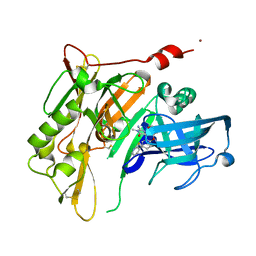 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | Descriptor: | (4S,4a'R,10a'S)-2-amino-8'-(2-fluoropyridin-3-yl)-1,4a'-dimethyl-3',4',4a',10a'-tetrahydro-2'H-spiro[imidazole-4,10'-pyrano[3,2-b]chromen]-5(1H)-one, Beta-secretase 1, NICKEL (II) ION | | Authors: | Thomas, A.A, Hunt, K.W, Newhouse, B, Watts, R.J, Liu, X, Vigers, G.P.A, Smith, D, Rhodes, S.P, Brown, K.D, Otten, J.N, Burkard, M, Cox, A.A, Geck Do, M.K, Dutcher, D, Rana, S, DeLisle, R.K, Regal, K, Wright, A.D, Groneberg, R, Liao, J, Scearce-Levie, K, Siu, M, Purkey, H.E, Lyssikatos, J.P. | | Deposit date: | 2014-11-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
8CF4
 
 | |
8CHG
 
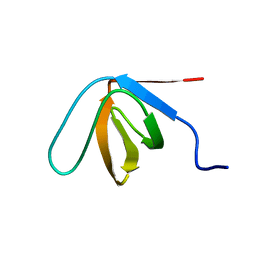 | |
8CE6
 
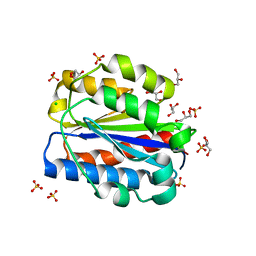 | | Crystal structure of human Cd11b I domain in P212121 space group | | Descriptor: | CHLORIDE ION, GLYCEROL, Integrin alpha-M, ... | | Authors: | Koekemoer, L, Williams, E, Ni, X, Gao, Q, Coker, J, MacLean, E.M, Wright, N.D, Marsden, B, Von Delft, F, Schwenzer, A, Midwood, K. | | Deposit date: | 2023-02-01 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | Crystal structure of human Cd11b I domain in P212121 space group
To Be Published
|
|
5FWB
 
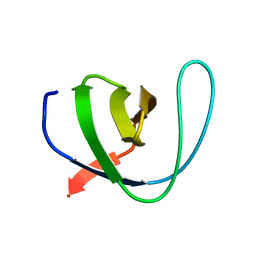 | | Human Spectrin SH3 domain D48G, E7F, K60F | | Descriptor: | SPECTRIN ALPHA CHAIN, NON-ERYTHROCYTIC 1 | | Authors: | Gallego, P, Navarro, S, Ventura, S, Reverter, D. | | Deposit date: | 2016-02-12 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Human Spectrin SH3 Domain D48G, E7F, K60F
To be Published
|
|
7Q89
 
 | | OleP mutant G92W in complex with 6DEB | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Vallone, B, Exertier, C, Freda, I, Gugole, E. | | Deposit date: | 2021-11-10 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Point Mutations at a Key Site Alter the Cytochrome P450 OleP Structural Dynamics.
Biomolecules, 12, 2021
|
|
5F81
 
 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Roessler, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2015-12-08 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
5F8X
 
 | | The crystal structure of human plasma kallikrein in complex with its peptide inhibitor pkalin-3 | | Descriptor: | CYS-PRO-ALA-ARG-PHE-M70-ALA-LEU-TRP-CYS, Plasma kallikrein, SULFATE ION, ... | | Authors: | Xu, M, Jiang, L, Xu, P, Luo, Z, Andreasen, P, Huang, M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure Of Human Plasma Kallikrein In Complex With Its Peptide Inhibitor Pkalin-3
To Be Published
|
|
4RRS
 
 | | 8-Tetrahydropyran-2-yl chromans: highly selective beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors | | Descriptor: | (4R,4a'R,10a'S)-8'-(2-fluoropyridin-3-yl)-4a'-methyl-3',4',4a',10a'-tetrahydro-2'H-spiro[1,3-oxazole-4,10'-pyrano[3,2-b]chromen]-2-amine, Beta-secretase 1, NICKEL (II) ION | | Authors: | Thomas, A.A, Hunt, K.W, Newhouse, B, Watts, R.J, Liu, X, Vigers, G.P.A, Smith, D, Rhodes, S.P, Brown, K.D, Otten, J.N, Burkard, M, Cox, A.A, Geck Do, M.K, Dutcher, D, Rana, S, DeLisle, R.K, Regal, K, Wright, A.D, Groneberg, R, Liao, J, Scearce-Levie, K, Siu, M, Purkey, H.E, Lyssikatos, J.P. | | Deposit date: | 2014-11-06 | | Release date: | 2014-12-03 | | Last modified: | 2014-12-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 8-Tetrahydropyran-2-yl Chromans: Highly Selective Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors.
J.Med.Chem., 57, 2014
|
|
4S30
 
 | |
7JW5
 
 | | Crystal structure of WT-CYP199A4 in complex with 4-phenylbenzoic acid | | Descriptor: | CHLORIDE ION, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2020-08-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.526 Å) | | Cite: | Different Geometric Requirements for Cytochrome P450-Catalyzed Aliphatic Versus Aromatic Hydroxylation Results in Chemoselective Oxidation
Acs Catalysis, 12, 2022
|
|
7Q6X
 
 | | OleP mutant S240Y in complex with 6DEB | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, FORMIC ACID, ... | | Authors: | Savino, C, Montemiglio, L.C, Vallone, B, Exertier, C, Freda, I, Gugole, E. | | Deposit date: | 2021-11-09 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Point Mutations at a Key Site Alter the Cytochrome P450 OleP Structural Dynamics.
Biomolecules, 12, 2021
|
|
4RSZ
 
 | |
