7TB8
 
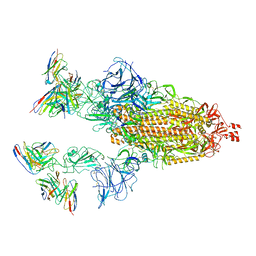 | | Cryo-EM structure of SARS-CoV-2 spike in complex with antibodies B1-182.1 and A19-61.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, T, Tsybovsky, T, Kwong, P.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529.
Science, 376, 2022
|
|
8XAB
 
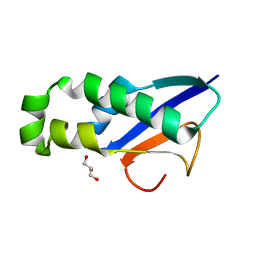 | |
5KVH
 
 | | Crystal structure of human apoptosis-inducing factor with W196A mutation | | Descriptor: | Apoptosis-inducing factor 1, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Brosey, C.A, Nix, J, Ellenberger, T, Tainer, J.A. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Defining NADH-Driven Allostery Regulating Apoptosis-Inducing Factor.
Structure, 24, 2016
|
|
7D2Z
 
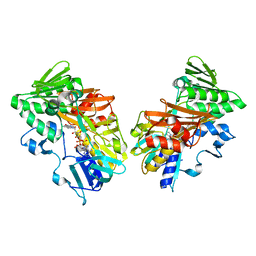
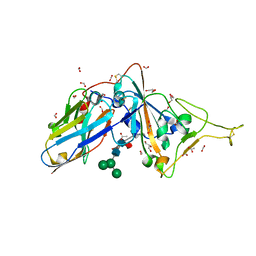 | | Structure of sybody SR31 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | ACETATE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A high-affinity RBD-targeting nanobody improves fusion partner's potency against SARS-CoV-2.
Plos Pathog., 17, 2021
|
|
7UJV
 
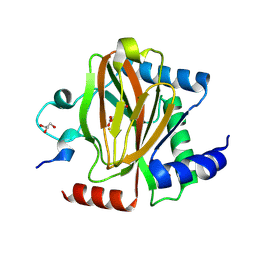 | | Structure of PHD2 in complex with HIF2a-CODD | | Descriptor: | Egl nine homolog 1, Endothelial PAS domain-containing protein 1, FE (III) ION, ... | | Authors: | Ferens, F.G, Tarade, D, Lee, J.E, Ohh, M. | | Deposit date: | 2022-03-31 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deficiency in PHD2-mediated hydroxylation of HIF2 alpha underlies Pacak-Zhuang syndrome.
Commun Biol, 7, 2024
|
|
7D30
 
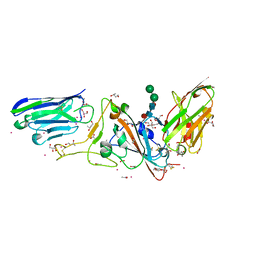 | | Structure of sybody MR17-SR31 fusion in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 1,3-BUTANEDIOL, ACETATE ION, CADMIUM ION, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A high-affinity RBD-targeting nanobody improves fusion partner's potency against SARS-CoV-2.
Plos Pathog., 17, 2021
|
|
5UA0
 
 | |
6YQ2
 
 | |
7T9H
 
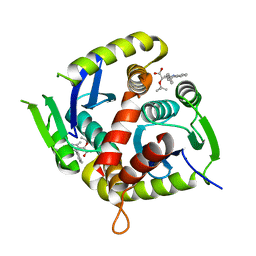 | | HIV Integrase in complex with Compound-15 | | Descriptor: | (2S)-tert-butoxy[2-methyl-4-(4-methylphenyl)quinolin-3-yl]acetic acid, Integrase, MAGNESIUM ION | | Authors: | Khan, J.A, Lewis, H, Kish, K. | | Deposit date: | 2021-12-19 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Design, Synthesis, and Preclinical Profiling of GSK3739936 (BMS-986180), an Allosteric Inhibitor of HIV-1 Integrase with Broad-Spectrum Activity toward 124/125 Polymorphs.
J.Med.Chem., 65, 2022
|
|
7T9O
 
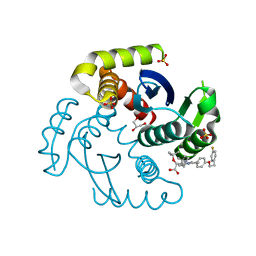 | | HIV Integrase in complex with Compound-25 | | Descriptor: | (2S)-tert-butoxy[4-(4,4-dimethylpiperidin-1-yl)-5-{4-[2-(4-fluorophenyl)ethoxy]phenyl}-2,6-dimethylpyridin-3-yl]acetic acid, GLYCEROL, Integrase, ... | | Authors: | Khan, J.A, Lewis, H, Kish, K. | | Deposit date: | 2021-12-19 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis, and Preclinical Profiling of GSK3739936 (BMS-986180), an Allosteric Inhibitor of HIV-1 Integrase with Broad-Spectrum Activity toward 124/125 Polymorphs.
J.Med.Chem., 65, 2022
|
|
7ZYT
 
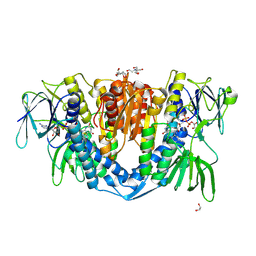 | | Crystal structure of the I318T pathogenic variant of the human dihydrolipoamide dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, ... | | Authors: | Nemes-Nikodem, E, Szabo, E, Vass, K.R, Lennartz, F, Nagy, B, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2022-05-25 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | Structural and Biochemical Investigation of Selected Pathogenic Mutants of the Human Dihydrolipoamide Dehydrogenase.
Int J Mol Sci, 24, 2023
|
|
8A6W
 
 | |
5TQO
 
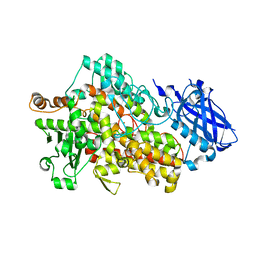 | | Lipoxygenase-1 (soybean) L546A/L754A mutant at 300K | | Descriptor: | FE (III) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Poss, E.M, Fraser, J.S, Gee, C. | | Deposit date: | 2016-10-24 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biophysical Characterization of a Disabled Double Mutant of Soybean Lipoxygenase: The "Undoing" of Precise Substrate Positioning Relative to Metal Cofactor and an Identified Dynamical Network.
J.Am.Chem.Soc., 141, 2019
|
|
7KJK
 
 | | The Neck region of Phage XM1 (6-fold symmetry) | | Descriptor: | Collar spike protein, Head completion protein, Tail sheath protein, ... | | Authors: | Wang, Z, Klose, T, Jiang, W, Kuhn, R.J. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of Vibrio phage XM1, a simple contractile DNA injection machine
Biorxiv, 2021
|
|
7KFA
 
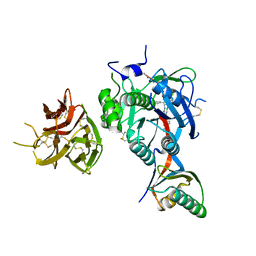 | | PCSK9 in complex with PCSK9i a 13mer cyclic peptide LDLR disruptor | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Chopra, R, Xu, M, Spraggon, G. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification of a PCSK9-LDLR disruptor peptide with in vivo function.
Cell Chem Biol, 29, 2022
|
|
7UOI
 
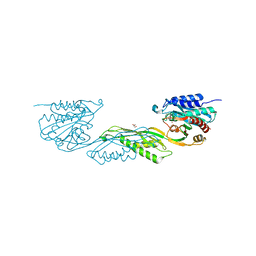 | | Crystallographic structure of DapE from Enterococcus faecium | | Descriptor: | GLYCEROL, ZINC ION, succinyl-diaminopimelate desuccinylase | | Authors: | Gonzalez-Segura, L, Diaz-Vilchis, A, Terrazas-Lopez, M, Diaz-Sanchez, A.G. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The three-dimensional structure of DapE from Enterococcus faecium reveals new insights into DapE/ArgE subfamily ligand specificity.
Int.J.Biol.Macromol., 270, 2024
|
|
6YPY
 
 | |
6YR8
 
 | | Affimer K6 - KRAS protein complex | | Descriptor: | Affimer K6, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Trinh, C.H, Haza, K.Z, Rao, A, Martin, H.L, Tiede, C, Edwards, T.A, McPherson, M.J, Tomlinson, D.C. | | Deposit date: | 2020-04-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RAS-inhibiting biologics identify and probe druggable pockets including an SII-alpha 3 allosteric site.
Nat Commun, 12, 2021
|
|
7ZZI
 
 | | Plasmodium falciparum hexokinase complexed with glucose and citrate | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Phosphotransferase, ... | | Authors: | Fritz-Wolf, K, Dillenberger, M, Rahlfs, S, Becker, K. | | Deposit date: | 2022-05-25 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of Plasmodium falciparum Hexokinase Provides Novel Information about Catalysis Due to a Plasmodium -Specific Insertion.
Int J Mol Sci, 24, 2023
|
|
6XUK
 
 | | AbLIFT design 15 of Ab 1116NS19.9 | | Descriptor: | GLYCEROL, Heavy chain, Light chain, ... | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
7T38
 
 | |
5AKC
 
 | | MutS in complex with the N-terminal domain of MutL - crystal form 2 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.6 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
5NPU
 
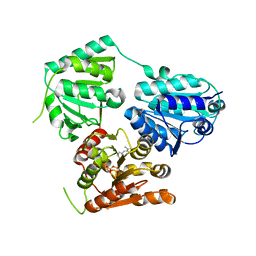 | | Inferred ancestral pyruvate decarboxylase | | Descriptor: | ANC27, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Buddrus, L, Crennell, S.J, Leak, D.J, Danson, M.J, Andrews, E.S.V, Arcus, V.L. | | Deposit date: | 2017-04-19 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of an inferred ancestral bacterial pyruvate decarboxylase.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7T37
 
 | | Activated state of 2-APB and CBD-bound wildtype rat TRPV2 in nanodiscs | | Descriptor: | 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 2, cannabidiol | | Authors: | Pumroy, R.A, Protopopova, A.D, Gallo, P.N, Moiseenkova-Bell, V.Y. | | Deposit date: | 2021-12-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into TRPV2 activation by small molecules.
Nat Commun, 13, 2022
|
|
5U5I
 
 | |
