6ZUA
 
 | | Cu nitrite reductase from Achromobacter cycloclastes: MSOX series at 170K, dose point 4 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Hough, M.A, Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2020-07-22 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Nature of the copper-nitrosyl intermediates of copper nitrite reductases during catalysis.
Chem Sci, 11, 2020
|
|
6ZUD
 
 | | Cu nitrite reductase from Achromobacter cycloclastes: MSOX series at 170K, dose point 3 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Hough, M.A, Antonyuk, S.V, Strange, R.W, Hasnain, S.S. | | Deposit date: | 2020-07-22 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Nature of the copper-nitrosyl intermediates of copper nitrite reductases during catalysis.
Chem Sci, 11, 2020
|
|
1DAH
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7,8-DIAMINO-NONANOIC ACID, 5'-ADENOSYL-METHYLENE-TRIPHOSPHATE, AND MANGANESE | | Descriptor: | 7,8-DIAMINO-NONANOIC ACID, DETHIOBIOTIN SYNTHETASE, MANGANESE (II) ION, ... | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
8ATC
 
 | | COMPLEX OF N-PHOSPHONACETYL-L-ASPARTATE WITH ASPARTATE CARBAMOYLTRANSFERASE. X-RAY REFINEMENT, ANALYSIS OF CONFORMATIONAL CHANGES AND CATALYTIC AND ALLOSTERIC MECHANISMS | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE (R STATE), CATALYTIC CHAIN, ASPARTATE CARBAMOYLTRANSFERASE REGULATORY CHAIN, ... | | Authors: | Ke, H, Lipscomb, W.N, Cho, Y, Honzatko, R.B. | | Deposit date: | 1989-08-25 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complex of N-phosphonacetyl-L-aspartate with aspartate carbamoyltransferase. X-ray refinement, analysis of conformational changes and catalytic and allosteric mechanisms.
J.Mol.Biol., 204, 1988
|
|
1DAI
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID | | Descriptor: | 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, DETHIOBIOTIN SYNTHETASE | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
4OJM
 
 | |
3OIK
 
 | | Human Carbonic anhydrase II mutant A65S, N67Q (CA IX mimic) bound by 2-Ethylestradiol 3,17-O,O-bis-sulfamate | | Descriptor: | (9BETA,13ALPHA,14BETA,17ALPHA)-2-ETHYLESTRA-1(10),2,4-TRIENE-3,17-DIYL DISULFAMATE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
257L
 
 | | AN ADAPTABLE METAL-BINDING SITE ENGINEERED INTO T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Wray, J.W, Baase, W.A, Ostheimer, G.J, Matthews, B.W. | | Deposit date: | 1999-01-05 | | Release date: | 2000-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Use of a non-rigid region in T4 lysozyme to design an adaptable metal-binding site.
Protein Eng., 13, 2000
|
|
8AJM
 
 | | Structure of human DDB1-DCAF12 in complex with the C-terminus of CCT5 | | Descriptor: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, T-complex protein 1 subunit epsilon | | Authors: | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | Deposit date: | 2022-07-28 | | Release date: | 2022-11-09 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8AJN
 
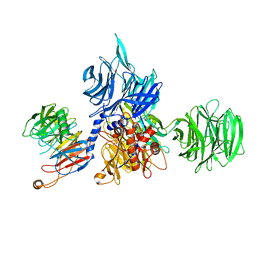 | | Structure of the human DDB1-DCAF12 complex | | Descriptor: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1 | | Authors: | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | Deposit date: | 2022-07-28 | | Release date: | 2022-11-09 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8AJO
 
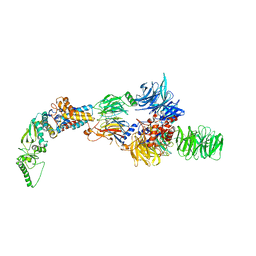 | | Negative-stain electron microscopy structure of DDB1-DCAF12-CCT5 | | Descriptor: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, T-complex protein 1 subunit epsilon | | Authors: | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | Deposit date: | 2022-07-28 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (30.6 Å) | | Cite: | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8QEZ
 
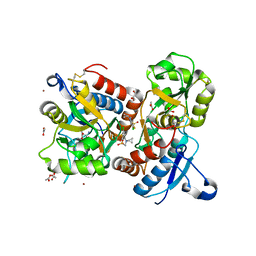 | | Crystal structure of the AMPA receptor GluA2-L504Y-N775S ligand binding domain in complex with L-glutamate and positive allosteric modulator BPAM395 at 1.55A resolution | | Descriptor: | 6-chloranyl-4-cyclopropyl-2,3-dihydrothieno[3,2-e][1,2,4]thiadiazine 1,1-dioxide, ACETATE ION, CACODYLATE ION, ... | | Authors: | Dorosz, J, Laulumaa, S, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2023-09-01 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Exploring thienothiadiazine dioxides as isosteric analogues of benzo- and pyridothiadiazine dioxides in the search of new AMPA and kainate receptor positive allosteric modulators.
Eur.J.Med.Chem., 264, 2023
|
|
3PFW
 
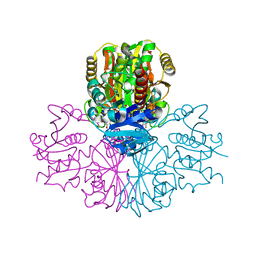 | | Crystal structure of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase (GAPDS) complex with NAD, a binary form | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, testis-specific, ... | | Authors: | Chaikuad, A, Shafqat, N, Yue, W.W, Cocking, R, Bray, J.E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and kinetic characterization of human sperm-specific glyceraldehyde-3-phosphate dehydrogenase, GAPDS.
Biochem.J., 435, 2011
|
|
8SS3
 
 | | Structure of LBD-TMD of AMPA receptor GluA2 in complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to competitive antagonist ZK and channel blocker spermidine (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, Digitonin, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SS2
 
 | | Structure of AMPA receptor GluA2 complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to competitive antagonist ZK and channel blocker spermidine (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, Digitonin, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6X3V
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus etomidate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Etomidate, ... | | Authors: | Kim, J.J, Gharpure, A, Teng, J, Zhuang, Y, Howard, R.J, Zhu, S, Noviello, C.M, Walsh, R.M, Lindahl, E, Hibbs, R.E. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Shared structural mechanisms of general anaesthetics and benzodiazepines.
Nature, 585, 2020
|
|
5XLF
 
 | | Crystal structure of aerobically purified and aerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
3QHM
 
 | | Crystal analysis of the complex structure, E342A-cellotetraose, of endocellulase from pyrococcus horikoshii | | Descriptor: | 458aa long hypothetical endo-1,4-beta-glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kim, H.-W, Ishikawa, K. | | Deposit date: | 2011-01-26 | | Release date: | 2012-02-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Functional analysis of hyperthermophilic endocellulase from Pyrococcus horikoshii by crystallographic snapshots
Biochem.J., 437, 2011
|
|
5XLG
 
 | | Crystal structure of anaerobically purified and aerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
5XLH
 
 | | Crystal structure of aerobically purified and aerobically crystallized for 12weeks D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
5XLE
 
 | | Crystal structure of anaerobically purified and anaerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
8AXD
 
 | | Human serotonin 5-HT3A receptor (apo, resting conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A | | Authors: | Lopez-Sanchez, U, Nury, H. | | Deposit date: | 2022-08-31 | | Release date: | 2024-03-06 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural determinants for activity of the antidepressant vortioxetine at human and rodent 5-HT 3 receptors.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QIC
 
 | |
1DAF
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, ADP, AND CALCIUM | | Descriptor: | 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
1DAE
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 3-(1-AMINOETHYL) NONANEDIOIC ACID | | Descriptor: | 3-(1-AMINOETHYL)NONANEDIOIC ACID, DETHIOBIOTIN SYNTHETASE | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
