4PA1
 
 | |
3MGC
 
 | | Teg12 Apo | | Descriptor: | GLYCEROL, IMIDAZOLE, N-L-ALPHA-ASPARTYL L-PHENYLALANINE 1-METHYL ESTER, ... | | Authors: | Bick, M.J, Banik, J.J, Darst, S.A, Brady, S.F. | | Deposit date: | 2010-04-05 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structures of the glycopeptide sulfotransferase Teg12 in a complex with the teicoplanin aglycone.
Biochemistry, 49, 2010
|
|
7V4G
 
 | | Crystal structure of human ALKBH5 in complex with m6A-containing ssRNA | | Descriptor: | GLYCEROL, MANGANESE (II) ION, RNA (5'-R(P*GP*GP*(6MZ)P*C)-3'), ... | | Authors: | Kaur, S, McDonough, M.A, Schofield, C.J, Aik, W.S. | | Deposit date: | 2021-08-13 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanisms of substrate recognition and N6-methyladenosine demethylation revealed by crystal structures of ALKBH5-RNA complexes.
Nucleic Acids Res., 50, 2022
|
|
3O2H
 
 | | E. coli ClpS in complex with a Leu N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS, DNA protection during starvation protein | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
1A05
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 3-ISOPROPYLMALATE DEHYDROGENASE FROM THIOBACILLUS FERROOXIDANS WITH 3-ISOPROPYLMALATE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, 3-ISOPROPYLMALIC ACID, MAGNESIUM ION | | Authors: | Imada, K, Inagaki, K, Matsunami, H, Kawaguchi, H, Tanaka, H, Tanaka, N, Namba, K. | | Deposit date: | 1997-12-09 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of 3-isopropylmalate dehydrogenase in complex with 3-isopropylmalate at 2.0 A resolution: the role of Glu88 in the unique substrate-recognition mechanism.
Structure, 6, 1998
|
|
4OGP
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P21) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA topoisomerase 2-associated protein PAT1 | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
1YY0
 
 | | Crystal structure of an RNA duplex containing a 2'-amine substitution and a 2'-amide product produced by in-crystal acylation at a C-A mismatch | | Descriptor: | 5'-R(*GP*CP*AP*GP*AP*(A5M)P*UP*UP*AP*AP*AP*UP*CP*UP*GP*C)-3', 5'-R(*GP*CP*AP*GP*AP*(M5M)P*UP*UP*AP*AP*AP*UP*CP*UP*GP*C)-3', CALCIUM ION | | Authors: | Gherghe, C.M, Krahn, J.M, Weeks, K.M. | | Deposit date: | 2005-02-23 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures, reactivity and inferred acylation transition States for 2'-amine substituted RNA.
J.Am.Chem.Soc., 127, 2005
|
|
1AOH
 
 | |
1Z79
 
 | |
3NLB
 
 | | Novel kinase profile highlights the temporal basis of context dependent checkpoint pathways to cell death | | Descriptor: | 3-methyl-5-[5-(1-methylethyl)-1H-benzimidazol-2-yl]-N-(1-methylpiperidin-4-yl)-1H-pyrazole-4-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Massey, A.J, Borgognoni, J, Bentley, C, Foloppe, N, Fiumana, A, Walmsley, L. | | Deposit date: | 2010-06-21 | | Release date: | 2011-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Context-dependent cell cycle checkpoint abrogation by a novel kinase inhibitor
Plos One, 5, 2010
|
|
1YZD
 
 | |
3N5E
 
 | | Crystal Structure of human thymidylate synthase bound to a peptide inhibitor | | Descriptor: | SULFATE ION, Synthetic peptide LR, Thymidylate synthase | | Authors: | Pozzi, C, Cardinale, D, Guaitoli, G, Tondi, D, Luciani, R, Myllykallio, H, Ferrari, S, Costi, M.P, Mangani, S. | | Deposit date: | 2010-05-25 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Protein-protein interface-binding peptides inhibit the cancer therapy target human thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1CQZ
 
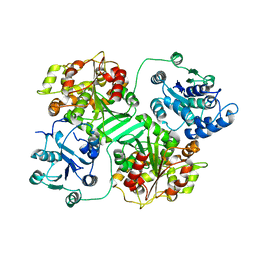 | | CRYSTAL STRUCTURE OF MURINE SOLUBLE EPOXIDE HYDROLASE. | | Descriptor: | EPOXIDE HYDROLASE | | Authors: | Argiriadi, M.A, Morisseau, C, Hammock, B.D, Christianson, D.W. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Detoxification of environmental mutagens and carcinogens: structure, mechanism, and evolution of liver epoxide hydrolase.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1AOP
 
 | |
1CSP
 
 | |
8EAP
 
 | |
7XGA
 
 | |
3NB3
 
 | | The host outer membrane proteins OmpA and OmpC are packed at specific sites in the Shigella phage Sf6 virion as structural components | | Descriptor: | Outer membrane protein A, Outer membrane protein C | | Authors: | Zhao, H, Sequeira, R.D, Galeva, N.A, Tang, L. | | Deposit date: | 2010-06-02 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | The host outer membrane proteins OmpA and OmpC are associated with the Shigella phage Sf6 virion.
Virology, 409, 2011
|
|
7VCL
 
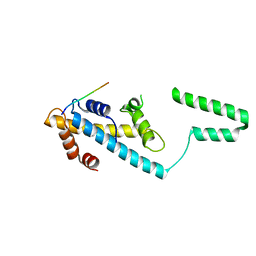 | |
3N5G
 
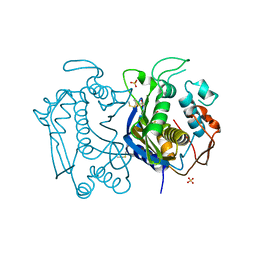 | | Crystal Structure of histidine-tagged human thymidylate synthase | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Pozzi, C, Cardinale, D, Guaitoli, G, Tondi, D, Luciani, R, Myllykallio, H, Ferrari, S, Costi, M.P, Mangani, S. | | Deposit date: | 2010-05-25 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Protein-protein interface-binding peptides inhibit the cancer therapy target human thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
268D
 
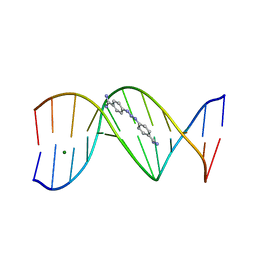 | |
4RV7
 
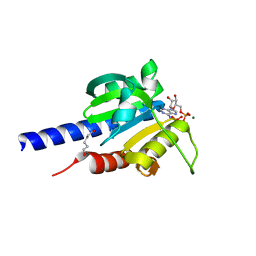 | | Characterization of an essential diadenylate cyclase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Diadenylate cyclase, HEXANE-1,6-DIOL, ... | | Authors: | Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2014-11-25 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Biochemical Analysis of the Essential Diadenylate Cyclase CdaA from Listeria monocytogenes.
J.Biol.Chem., 290, 2015
|
|
269D
 
 | | STRUCTURAL STUDIES ON NUCLEIC ACIDS | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(5CM)P*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Partridge, B.L, Salisbury, S.A. | | Deposit date: | 1996-07-12 | | Release date: | 1996-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: |
To be Published, 1996
|
|
1CSQ
 
 | |
5C9H
 
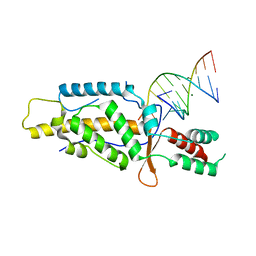 | | Structural Basis of Template Boundary Definition in Tetrahymena Telomerase | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*AP*GP*AP*AP*CP*UP*GP*UP*CP*A)-3'), RNA (5'-R(P*UP*CP*AP*UP*UP*CP*AP*GP*UP*UP*CP*U)-3'), ... | | Authors: | Jansson, L.I, Akiyama, B.M, Ooms, A, Lu, C, Rubin, S.M, Stone, M.D. | | Deposit date: | 2015-06-26 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of template-boundary definition in Tetrahymena telomerase.
Nat.Struct.Mol.Biol., 22, 2015
|
|
