1B3Y
 
 | | XYLANASE FROM PENICILLIUM SIMPLICISSIMUM, COMPLEX WITH XYLOTETRAOSE | | Descriptor: | PROTEIN (XYLANASE), alpha-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Schmidt, A, Kratky, C. | | Deposit date: | 1998-12-15 | | Release date: | 1999-04-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Xylan binding subsite mapping in the xylanase from Penicillium simplicissimum using xylooligosaccharides as cryo-protectant.
Biochemistry, 38, 1999
|
|
4UMJ
 
 | | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01, with bound ibandronic acid molecules. | | Descriptor: | GERANYLTRANSTRANSFERASE, IBANDRONATE, MAGNESIUM ION | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2014-05-18 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Characterization of Substrate and Inhibitor Binding to Farnesyl Pyrophosphate Synthase from Pseudomonas Aeruginosa.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7NMI
 
 | |
7NEM
 
 | | Hydrogenase-2 variant R479K - anaerobically oxidised form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B. | | Deposit date: | 2021-02-04 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A comprehensive structural and kinetic investigation of the role of the active-site argininein bidirectional hydrogen activation by the [NiFe]-hydrogenase "Hyd-2) from Escherichia coli
To Be Published
|
|
4UIU
 
 | | BROMODOMAIN OF HUMAN BRD9 WITH 7-(3,4-dimethoxyphenyl)-N-(1,1-dioxo-1- thian-4-yl)-5-methyl-4-oxo-4H,5H-thieno-3,2-c-pyridine-2-carboxamide | | Descriptor: | BROMODOMAIN-CONTAINING PROTEIN 9, N-[1,1-bis(oxidanylidene)thian-4-yl]-7-(3,4-dimethoxyphenyl)-5-methyl-4-oxidanylidene-thieno[3,2-c]pyridine-2-carboxamide | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
4ULW
 
 | |
4UFY
 
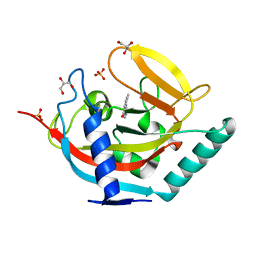 | | Crystal structure of human tankyrase 2 in complex with TA-13 | | Descriptor: | 8-METHYL-2-(4-METHYLPHENYL)-3,4-DIHYDROQUINAZOLIN-4-ONE, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-20 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
4UDX
 
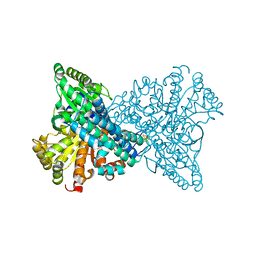 | | CO2 bound to cluster C of Ni,Fe-CO dehydrogenase at true-atomic resolution | | Descriptor: | CARBON DIOXIDE, CARBON MONOXIDE DEHYDROGENASE 2, FE (II) ION, ... | | Authors: | Fesseler, J, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2014-12-12 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | How the [NiFe4S4] Cluster of CO Dehydrogenase Activates CO2 and NCO(-).
Angew. Chem. Int. Ed. Engl., 54, 2015
|
|
4UBL
 
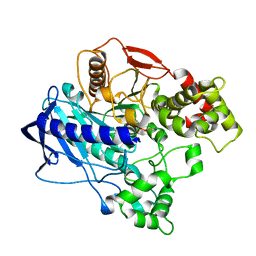 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 9.26 MGy | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-13 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
3HII
 
 | |
1AVB
 
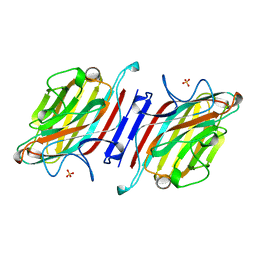 | | ARCELIN-1 FROM PHASEOLUS VULGARIS L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARCELIN-1, ... | | Authors: | Mourey, L, Pedelacq, J.D, Fabre, C, Rouge, P, Samama, J.P. | | Deposit date: | 1997-09-15 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the arcelin-1 dimer from Phaseolus vulgaris at 1.9-A resolution.
J.Biol.Chem., 273, 1998
|
|
7N0E
 
 | |
1AXM
 
 | | HEPARIN-LINKED BIOLOGICALLY-ACTIVE DIMER OF FIBROBLAST GROWTH FACTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | DiGabriele, A.D, Lax, I, Chen, D.I, Svahn, C.M, Jaye, M, Schlessinger, J, Hendrickson, W.A. | | Deposit date: | 1997-10-16 | | Release date: | 1998-04-22 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a heparin-linked biologically active dimer of fibroblast growth factor.
Nature, 393, 1998
|
|
7NPA
 
 | |
4UI8
 
 | | Crystal structure of human tankyrase 2 in complex with TA-55 | | Descriptor: | 8-HYDROXY-2-[4-(TRIFLUOROMETHYL)PHENYL]-3,4-DIHYDROQUINAZOLIN-4-ONE, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
4UHI
 
 | | HUMAN STEROL 14-ALPHA DEMETHYLASE (CYP51) IN COMPLEX WITH VFV IN C121 SPACE GROUP | | Descriptor: | N-[(1R)-1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Hargrove, T.Y, Wawrzak, Z, I Lepesheva, G. | | Deposit date: | 2015-03-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Human Sterol 14Alpha-Demethylase (Cyp51) as a Target for Anticancer Chemotherapy: Towards Structure-Aided Drug Design.
J.Lipid Res., 57, 2016
|
|
7NNJ
 
 | | Crystal Structure of NUDT4 (Diphosphoinositol polyphosphate phosphohydrolase 2) in complex with 4-O-Bn-1-PCP-InsP4 (AMR2105) | | Descriptor: | 1,2-ETHANEDIOL, Diphosphoinositol polyphosphate phosphohydrolase 2, FORMIC ACID, ... | | Authors: | Dubianok, Y, Arruda Bezerra, G, Raux, B, Diaz Saez, L, Riley, A.M, Potter, B.V.L, Huber, K.V.M, von Delft, F. | | Deposit date: | 2021-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Crystal Structure of NUDT4 (Diphosphoinositol polyphosphate phosphohydrolase 2) in complex with 4-O-Bn-1-PCP-InsP4 (AMR2105)
To Be Published
|
|
4UCT
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-amino-6-methyl-5-(propan-2-yloxy)-3H-[1,2,4]triazolo[1,5-a]pyrimidin-8-ium, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UI7
 
 | | Crystal structure of human tankyrase 2 in complex with TA-49 | | Descriptor: | 8-HYDROXY-2-(4-METHYLPHENYL)-3,4-DIHYDROQUINAZOLIN-4-ONE, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
7NP8
 
 | |
7MYV
 
 | | Plasmodium falciparum HAD5/PMM | | Descriptor: | MAGNESIUM ION, Phosphomannomutase | | Authors: | Frasse, P.M, Odom John, A.R, Goldberg, D.E. | | Deposit date: | 2021-05-21 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Enzymatic and structural characterization of HAD5, an essential phosphomannomutase of malaria-causing parasites.
J.Biol.Chem., 298, 2022
|
|
7ZY9
 
 | |
4UB9
 
 | | Structural and catalytic characterization of molinate hydrolase | | Descriptor: | Molinate hydrolase, ZINC ION | | Authors: | Leite, J.P, Duarte, M, Paiva, A, Ferreira-da-Silva, F, Matias, P.M, Nunes, O, Gales, L. | | Deposit date: | 2014-08-12 | | Release date: | 2015-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-guided engineering of molinate hydrolase for the degradation of thiocarbamate pesticides.
Plos One, 10, 2015
|
|
7N3S
 
 | |
7N3R
 
 | |
