2F3S
 
 | |
1QMU
 
 | | Duck carboxypeptidase D domain II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBOXYPEPTIDASE GP180 RESIDUES 503-882, SULFATE ION, ... | | Authors: | Gomis-Rueth, F.X, Coll, M, Aviles, F.X, Vendrell, J, Fricker, L.D. | | Deposit date: | 1999-10-06 | | Release date: | 2000-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Avian Carboxypeptidase D Domain II : A Prototype for the Regulatory Metallocarboxypeptidase Subfamily
Embo J., 18, 1999
|
|
2F3Q
 
 | |
2F3P
 
 | |
2FFR
 
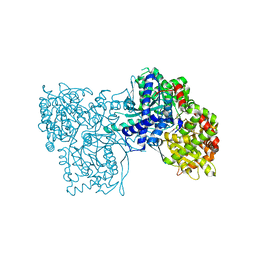 | | Crystallographic studies on N-azido-beta-D-glucopyranosylamine, an inhibitor of glycogen phosphorylase: comparison with N-acetyl-beta-D-glucopyranosylamine | | Descriptor: | Glycogen phosphorylase, muscle form, N-(azidoacetyl)-beta-D-glucopyranosylamine, ... | | Authors: | Petsalakis, E.I, Chrysina, E.D, Tiraidis, C, Hadjiloi, T, Leonidas, D.D, Oikonomakos, N.G, Aich, U, Varghese, B, Loganathan, D. | | Deposit date: | 2005-12-20 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystallographic studies on N-azidoacetyl-beta-d-glucopyranosylamine, an inhibitor of glycogen phosphorylase: Comparison with N-acetyl-beta-d-glucopyranosylamine.
Bioorg.Med.Chem., 14, 2006
|
|
6G1F
 
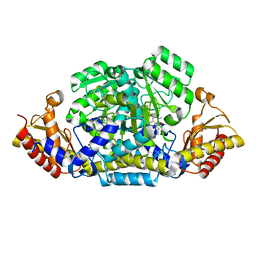 | |
8TM9
 
 | | Computationally designed tunable C2 symmetric tandem repeat homodimer, D_3_633_8x bound to peptide | | Descriptor: | D_3_633_8x peptide bound, MC1, N-PROPANOL, ... | | Authors: | Kennedy, M.A, Stoddard, B.L, Hicks, D.R. | | Deposit date: | 2023-07-28 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Computationally designed tunable C2 symmetric tandem repeat homodimer, D_3_633_8x bound to peptide
To Be Published
|
|
4Y4V
 
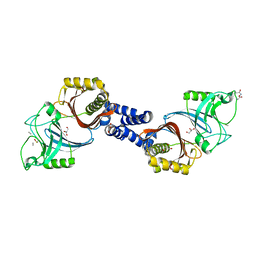 | | Structure of Helicobacter pylori Csd6 in the D-Ala-bound state | | Descriptor: | Conserved hypothetical secreted protein, D-ALANINE, GLYCEROL | | Authors: | Kim, H.S, Im, H.N, Yoon, H.J, Suh, S.W. | | Deposit date: | 2015-02-11 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Cell Shape-determining Csd6 Protein from Helicobacter pylori Constitutes a New Family of l,d-Carboxypeptidase
J.Biol.Chem., 290, 2015
|
|
5L34
 
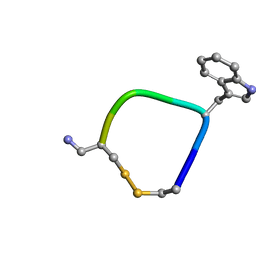 | |
6CKG
 
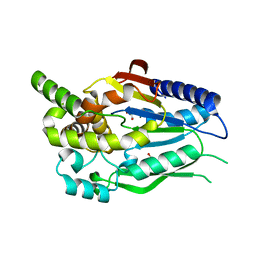 | | D-glycerate 3-kinase from Cryptococcus neoformans var. grubii serotype A (H99 / ATCC 208821 / CBS 10515 / FGSC 9487) | | Descriptor: | 1,2-ETHANEDIOL, D-glycerate 3-kinase | | Authors: | Horanyi, P.S, Abendroth, J, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-02-28 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D-glycerate 3-kinase from Cryptococcus neoformans var. grubii serotype A (H99 / ATCC 208821 / CBS 10515 / FGSC 9487)
To be Published
|
|
1ALZ
 
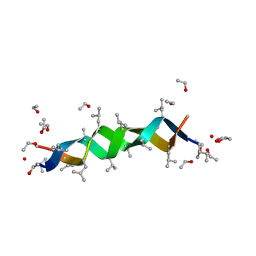 | | GRAMICIDIN D FROM BACILLUS BREVIS (ETHANOL SOLVATE) | | Descriptor: | ETHANOL, ILE-GRAMICIDIN C, VAL-GRAMICIDIN A | | Authors: | Burkhart, B.M, Pangborn, W.A, Duax, W.L, Langs, D.A. | | Deposit date: | 1997-06-06 | | Release date: | 1998-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Heterodimer Formation and Crystal Nucleation of Gramicidin D
Biophys.J., 75, 1998
|
|
432D
 
 | | D(GGCCAATTGG) COMPLEXED WITH DAPI | | Descriptor: | 6-AMIDINE-2-(4-AMIDINO-PHENYL)INDOLE, DNA (5'-D(*GP*GP*CP*CP*AP*AP*TP*TP*GP*G)-3') | | Authors: | Vlieghe, D, Van Meervelt, L. | | Deposit date: | 1998-10-14 | | Release date: | 1999-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of d(GGCCAATTGG) complexed with DAPI reveals novel binding mode.
Biochemistry, 38, 1999
|
|
1ALX
 
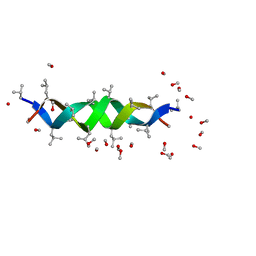 | | GRAMICIDIN D FROM BACILLUS BREVIS (METHANOL SOLVATE) | | Descriptor: | GRAMICIDIN A, METHANOL | | Authors: | Burkhart, B.M, Langs, D.A, Smith, G.D, Courseille, C, Precigoux, G, Hospital, M, Pangborn, W.A, Duax, W.L. | | Deposit date: | 1997-06-05 | | Release date: | 1998-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Heterodimer Formation and Crystal Nucleation of Gramicidin D
Biophys.J., 75, 1998
|
|
2XPC
 
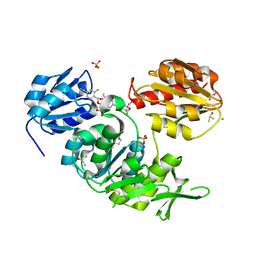 | | Second-generation sulfonamide inhibitors of MurD: Activity optimisation with conformationally rigid analogues of D-glutamic acid | | Descriptor: | (1R,3R,4S)-4-[({6-[(4-CYANO-2-FLUOROBENZYL)OXY]NAPHTHALEN-2-YL}SULFONYL)AMINO]CYCLOHEXANE-1,3-DICARBOXYLIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Sosic, I, Barreteau, H, Simcic, M, Sink, R, Cesar, J, Golic-Grdadolnik, S, Contreras-Martel, C, Dessen, A, Amoroso, A, Joris, B, Blanot, D, Gobec, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Second-Generation Sulfonamide Inhibitors of D- Glutamic Acid-Adding Enzyme: Activity Optimisation with Conformationally Rigid Analogues of D- Glutamic Acid.
Eur.J.Med.Chem, 46, 2011
|
|
7KJN
 
 | |
1ZUE
 
 | | Revised Solution Structure of DLP-2 | | Descriptor: | Defensin-like peptide 2/4 | | Authors: | Torres, A.M, Tsampazi, C, Geraghty, D.P, Bansal, P.S, Alewood, P.F, Kuchel, P.W. | | Deposit date: | 2005-05-30 | | Release date: | 2005-08-02 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | D-amino acid residue in a defensin-like peptide from platypus venom: effect on structure and chromatographic properties.
Biochem.J., 391, 2005
|
|
4Q3F
 
 | | Human D-DT complexed with tartrate | | Descriptor: | D-dopachrome decarboxylase, L(+)-TARTARIC ACID | | Authors: | Rajasekaran, D, Lolis, E. | | Deposit date: | 2014-04-11 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting distinct tautomerase sites of D-DT and MIF with a single molecule for inhibition of neutrophil lung recruitment.
Faseb J., 28, 2014
|
|
7U9K
 
 | | Staphylococcus aureus D-alanine-D-alanine ligase in complex with ATP, D-ala-D-ala, Mg2+ and K+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, D-alanine--D-alanine ligase, ... | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided design and synthesis of ATP-competitive N-acyl-substituted sulfamide d-alanine-d-alanine ligase inhibitors.
Bioorg.Med.Chem., 96, 2023
|
|
1UNO
 
 | | Crystal structure of a d,l-alternating peptide | | Descriptor: | H-(L-TYR-D-TYR)4-LYS-OH | | Authors: | Alexopoulos, E, Kuesel, A, Uson, I, Diederichsen, U, Sheldrick, G.M. | | Deposit date: | 2003-09-11 | | Release date: | 2004-09-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Solution and Structure of an Alternating D,L-Peptide
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1BQG
 
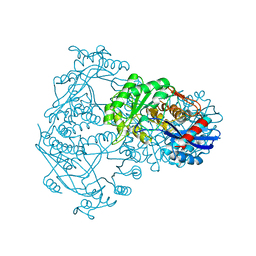 | | THE STRUCTURE OF THE D-GLUCARATE DEHYDRATASE PROTEIN FROM PSEUDOMONAS PUTIDA | | Descriptor: | D-GLUCARATE DEHYDRATASE | | Authors: | Gulick, A.M, Palmer, D.R.J, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | Deposit date: | 1998-08-15 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystal structure of (D)-glucarate dehydratase from Pseudomonas putida.
Biochemistry, 37, 1998
|
|
1XIB
 
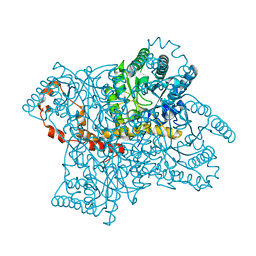 | |
3L8L
 
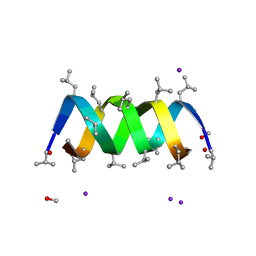 | | Gramicidin D complex with sodium iodide | | Descriptor: | GRAMICIDIN D, IODIDE ION, METHANOL, ... | | Authors: | Olczak, A, Glowka, M.L, Szczesio, M, Bojarska, J, Wawrzak, Z, Duax, W.L. | | Deposit date: | 2009-12-31 | | Release date: | 2010-07-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The first crystal structure of a gramicidin complex with sodium: high-resolution study of a nonstoichiometric gramicidin D-NaI complex.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1XLE
 
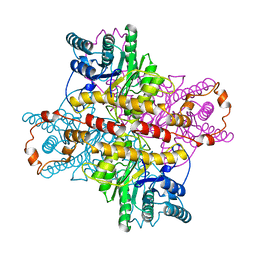 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, MANGANESE (II) ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLB
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLK
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, MANGANESE (II) ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
