6PFJ
 
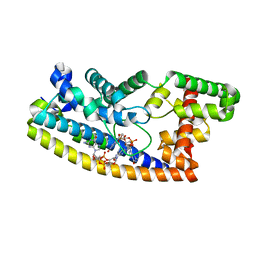 | | Structure of S. venezuelae RsiG-WhiG-(ci-di-GMP) complex, P64 crystal form | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), AmfC protein, RNA polymerase sigma factor | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-06-21 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | c-di-GMP Arms an Anti-sigma to Control Progression of Multicellular Differentiation in Streptomyces.
Mol.Cell, 77, 2020
|
|
6PFV
 
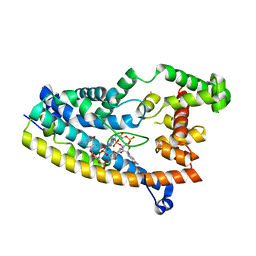 | | Structure of S. venezuelae RisG-WhiG-c-di-GMP complex: orthorhombic crystal form | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), AmfC protein, RNA polymerase sigma factor | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-06-22 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | c-di-GMP Arms an Anti-sigma to Control Progression of Multicellular Differentiation in Streptomyces.
Mol.Cell, 77, 2020
|
|
6IDO
 
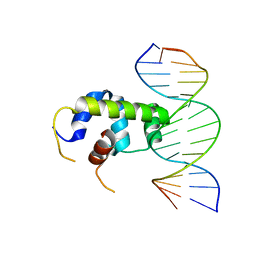 | | Crystal structure of Klebsiella pneumoniae sigma4 of sigmaS fusing with the RNA polymerase beta-flap-tip-helix in complex with -35 element DNA | | Descriptor: | DNA (5'-D(P*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*AP*AP*TP*CP*G)-3'), DNA (5'-D(P*GP*AP*TP*TP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*C)-3'), RNA polymerase sigma factor RpoS,RNA polymerase beta-flap-tip-helix | | Authors: | Lou, Y.C, Chien, C.Y, Chen, C, Hsu, C.H. | | Deposit date: | 2018-09-10 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.748 Å) | | Cite: | Structural basis for -35 element recognition by sigma4chimera proteins and their interactions with PmrA response regulator.
Proteins, 88, 2020
|
|
6OMF
 
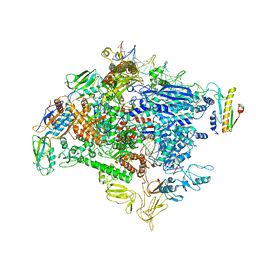 | | CryoEM structure of SigmaS-transcription initiation complex with activator Crl | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Jaramillo Cartagena, A, Darst, S.A, Campbell, E.A. | | Deposit date: | 2019-04-18 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for transcription activation by Crl through tethering of sigmaSand RNA polymerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K4Y
 
 | | CryoEM structure of sigma appropriation complex | | Descriptor: | 10 kDa anti-sigma factor, DNA (60-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structural basis of sigma appropriation.
Nucleic Acids Res., 47, 2019
|
|
6OY7
 
 | |
6OW3
 
 | |
6OVR
 
 | |
6OVY
 
 | |
6P71
 
 | |
6P18
 
 | | Q21 transcription antitermination complex: loading complex | | Descriptor: | DNA (67-MER) fragment carrying phage-21 pR' promoter and pause element, nontemplate strand, template strand, ... | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of Q-dependent antitermination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OY6
 
 | |
6P70
 
 | | X-ray crystal structure of bacterial RNA polymerase and pyrBI promoter complex | | Descriptor: | DNA (5'-D(*TP*AP*TP*AP*AP*TP*CP*GP*AP*TP*CP*TP*TP*TP*GP*CP*CP*GP*GP*G)-3'), DNA (5'-D(P*TP*CP*CP*CP*GP*GP*CP*AP*AP*AP*TP*TP*GP*TP*CP*CP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shin, Y, Murakami, K.S. | | Deposit date: | 2019-06-04 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Structural basis of reiterative transcription from the pyrG and pyrBI promoters by bacterial RNA polymerase.
Nucleic Acids Res., 48, 2020
|
|
6JNX
 
 | | Cryo-EM structure of a Q-engaged arrested complex | | Descriptor: | Antiterminator Q protein, DNA (63-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Feng, Y, Shi, J. | | Deposit date: | 2019-03-18 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis of Q-dependent transcription antitermination.
Nat Commun, 10, 2019
|
|
6OY5
 
 | |
6DVC
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF sigma factor L) containing 5nt RNA with 6nt spacer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2018-06-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
6DVB
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF sigma factor L) containing 5nt RNA with 5nt spacer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2018-06-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
6DV9
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF sigma factor L) containing 5nt RNA with 4nt spacer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*CP*AP*GP*AP*GP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2018-06-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
6DVE
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF selenomethionine-labelled sigma factor L) with 6 nt spacer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*T)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*GP*AP*GP*TP*AP*AP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2018-06-23 | | Release date: | 2019-02-20 | | Last modified: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (3.812 Å) | | Cite: | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
6DVD
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF sigma factor L) with 6 nt spacer and bromine labelled in position "-11 | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*A)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2018-06-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.899 Å) | | Cite: | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
6N61
 
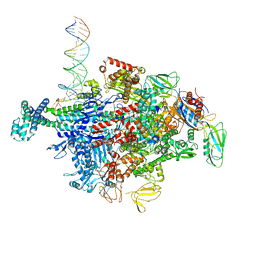 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA and Capistruin | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Capistruin, ... | | Authors: | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.253 Å) | | Cite: | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6N60
 
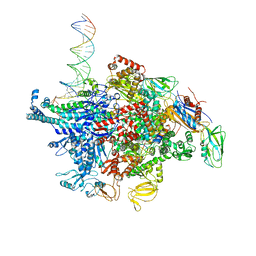 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA and Microcin J25 (MccJ25) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-23 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6N62
 
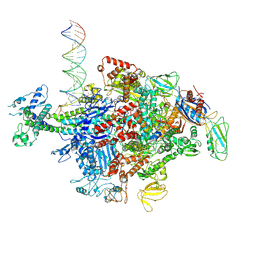 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.803 Å) | | Cite: | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6EYD
 
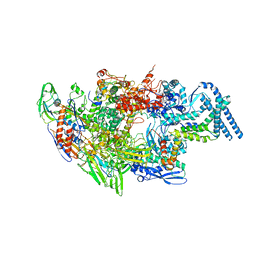 | | Structure of Mycobacterium smegmatis RNA polymerase Sigma-A holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Kouba, T, Barvik, I, Krasny, L. | | Deposit date: | 2017-11-11 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | The Core and Holoenzyme Forms of RNA Polymerase fromMycobacterium smegmatis.
J. Bacteriol., 201, 2019
|
|
6EEC
 
 | | Mycobacterium tuberculosis RNAP promoter unwinding intermediate complex with RbpA/CarD and AP3 promoter captured by Corallopyronin | | Descriptor: | DNA (63-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2018-08-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Nature, 565, 2019
|
|
