7Z12
 
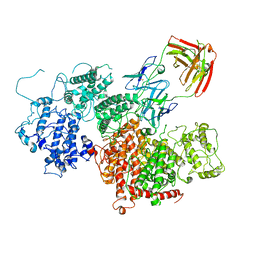 | | VAR2 complex with PAM1.4 | | Descriptor: | PAM1.4, Heavy Chain, light Chain, ... | | Authors: | Raghavan, S.S.R, Wang, K.T. | | Deposit date: | 2022-02-24 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM reveals the conformational epitope of human monoclonal antibody PAM1.4 broadly reacting with polymorphic malarial protein VAR2CSA.
Plos Pathog., 18, 2022
|
|
2WGH
 
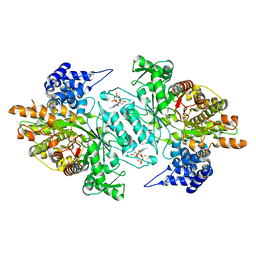 | | Human Ribonucleotide reductase R1 subunit (RRM1) in complex with dATP and Mg. | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Welin, R.M, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kragh-Nielsen, T, Kotzsch, A, Kotenyova, T, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Wisniewska, M, Nordlund, P. | | Deposit date: | 2009-04-19 | | Release date: | 2009-05-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Allosteric Regulation of Human Ribonucleotide Reductase by Nucleotide-Induced Oligomerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7YXA
 
 | | XFEL crystal structure of the human sphingosine 1 phosphate receptor 5 in complex with ONO-5430608 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[6-(2-naphthalen-1-ylethoxy)-2,3,4,5-tetrahydro-1H-3-benzazepin-3-ium-3-yl]butanoic acid, ... | | Authors: | Lyapina, E, Marin, E, Gusach, A, Orekhov, P, Gerasimov, A, Luginina, A, Vakhrameev, D, Ergasheva, M, Kovaleva, M, Khusainov, G, Khorn, P, Shevtsov, M, Kovalev, K, Okhrimenko, I, Bukhdruker, S, Popov, P, Hu, H, Weierstall, U, Liu, W, Cho, Y, Gushchin, I, Rogachev, A, Bourenkov, G, Park, S, Park, G, Huyn, H.J, Park, J, Gordeliy, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for receptor selectivity and inverse agonism in S1P 5 receptors.
Nat Commun, 13, 2022
|
|
7Z20
 
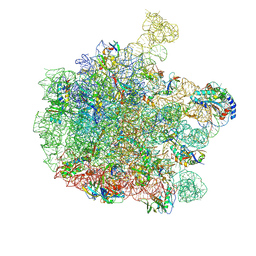 | | 70S E. coli ribosome with an extended uL23 loop from Candidatus marinimicrobia and a stalled filamin domain 5 nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Sidhu, H, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-02-25 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
1CD1
 
 | |
7YZA
 
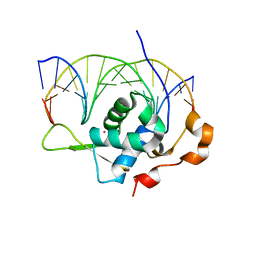 | | Crystal structure of the zebrafish FoxH1 bound to the TGTGTATT site | | Descriptor: | DNA (5'-D(*AP*GP*AP*TP*TP*GP*TP*GP*TP*AP*TP*TP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*CP*AP*AP*TP*AP*CP*AP*CP*AP*AP*TP*CP*T)-3'), Forkhead box protein H1, ... | | Authors: | Pluta, R, Macias, M.J. | | Deposit date: | 2022-02-19 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Molecular basis for DNA recognition by the maternal pioneer transcription factor FoxH1.
Nat Commun, 13, 2022
|
|
1BRD
 
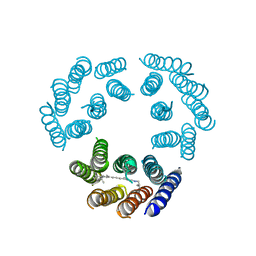 | | Model for the structure of Bacteriorhodopsin based on high-resolution Electron Cryo-microscopy | | Descriptor: | BACTERIORHODOPSIN PRECURSOR, RETINAL | | Authors: | Henderson, R, Baldwin, J.M, Ceska, T.A, Zemlin, F, Beckmann, E, Downing, K.H. | | Deposit date: | 1990-05-23 | | Release date: | 1991-04-15 | | Last modified: | 2024-04-17 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy.
J.Mol.Biol., 213, 1990
|
|
1BJN
 
 | |
3AQD
 
 | |
1CDK
 
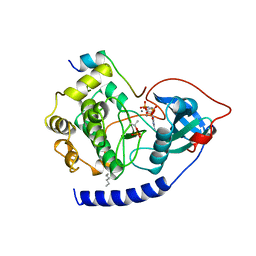 | | CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT (E.C.2.7.1.37) (PROTEIN KINASE A) COMPLEXED WITH PROTEIN KINASE INHIBITOR PEPTIDE FRAGMENT 5-24 (PKI(5-24) ISOELECTRIC VARIANT CA) AND MN2+ ADENYLYL IMIDODIPHOSPHATE (MNAMP-PNP) AT PH 5.6 AND 7C AND 4C | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, MANGANESE (II) ION, MYRISTIC ACID, ... | | Authors: | Bossemeyer, D, Engh, R.A, Kinzel, V, Ponstingl, H, Huber, R. | | Deposit date: | 1994-07-04 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 A structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5-24).
EMBO J., 12, 1993
|
|
1BJW
 
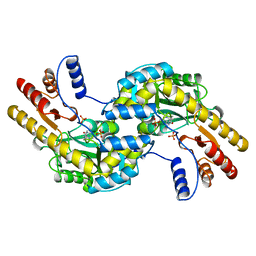 | | ASPARTATE AMINOTRANSFERASE FROM THERMUS THERMOPHILUS | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PHOSPHATE ION | | Authors: | Nakai, T, Okada, K, Kuramitsu, S, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-06-30 | | Release date: | 1999-07-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Thermus thermophilus HB8 aspartate aminotransferase and its complex with maleate.
Biochemistry, 38, 1999
|
|
2VSU
 
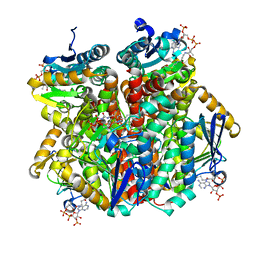 | | A ternary complex of Hydroxycinnamoyl-CoA Hydratase-Lyase (HCHL) with acetyl-Coenzyme A and vanillin gives insights into substrate specificity and mechanism. | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, ACETYL COENZYME *A, P-HYDROXYCINNAMOYL COA HYDRATASE/LYASE | | Authors: | Bennett, J.P, Bertin, L.M, Brzozowski, A.M, Walton, N.J, Grogan, G. | | Deposit date: | 2008-04-29 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Ternary Complex of Hydroxycinnamoyl-Coa Hydratase-Lyase (Hchl) with Acetyl-Coa and Vanillin Gives Insights Into Substrate Specificity and Mechanism.
Biochem.J., 414, 2008
|
|
2VSS
 
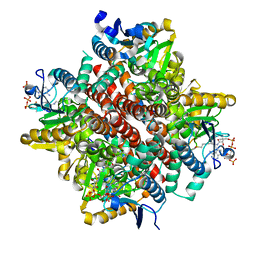 | | Wild-type Hydroxycinnamoyl-CoA hydratase lyase in complex with acetyl- CoA and vanillin | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, ACETYL COENZYME *A, P-HYDROXYCINNAMOYL COA HYDRATASE/LYASE | | Authors: | Bennett, J.P, Bertin, L.M, Brzozowski, A.M, Walton, N.J, Grogan, G. | | Deposit date: | 2008-04-29 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A Ternary Complex of Hydroxycinnamoyl-Coa Hydratase-Lyase (Hchl) with Acetyl-Coa and Vanillin Gives Insights Into Substrate Specificity and Mechanism.
Biochem.J., 414, 2008
|
|
2VQ3
 
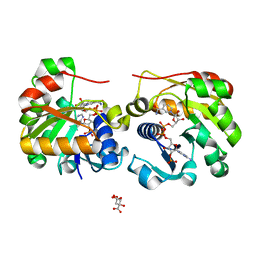 | | Crystal Structure of the Membrane Proximal Oxidoreductase Domain of Human Steap3, the Dominant Ferric Reductase of the Erythroid Transferrin Cycle | | Descriptor: | CITRIC ACID, METALLOREDUCTASE STEAP3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sendamarai, A.K, Ohgami, R.S, Fleming, M.D, Lawrence, C.M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-05-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Membrane Proximal Oxidoreductase Domain of Human Steap3, the Dominant Ferrireductase of the Erythroid Transferrin Cycle
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1CJC
 
 | |
8QZC
 
 | |
1CGX
 
 | |
8QPN
 
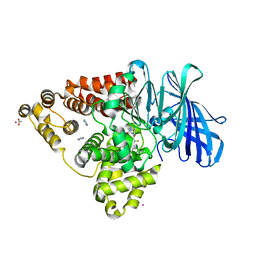 | | LTA4 hydrolase in complex with compound 6(S) | | Descriptor: | (2S)-2-azanyl-3-[5-[4-(5-chloranyl-3-fluoranyl-pyridin-2-yl)oxyphenyl]-1,2,3,4-tetrazol-2-yl]propan-1-ol, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2023-10-02 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Amino Alcohols as Highly Potent, Selective, and Orally Efficacious Inhibitors of Leukotriene A4 Hydrolase.
J.Med.Chem., 66, 2023
|
|
2WIF
 
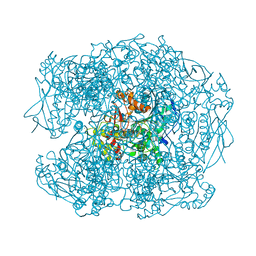 | | AGED FORM OF HUMAN BUTYRYLCHOLINESTERASE INHIBITED BY TABUN ANALOGUE TA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Carletti, E, Aurbek, N, Gillon, E, Loiodice, M, Nicolet, Y, Fontecilla, J, Masson, P, Thiermann, H, Nachon, F, Worek, F. | | Deposit date: | 2009-05-11 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-activity analysis of aging and reactivation of human butyrylcholinesterase inhibited by analogues of tabun.
Biochem. J., 421, 2009
|
|
8QOW
 
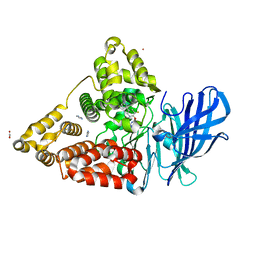 | | LTA4 hydrolase in complex with compound 2(S) | | Descriptor: | (2~{S})-2-azanyl-3-[3-[4-[3-fluoranyl-5-(1~{H}-pyrazol-5-yl)pyridin-2-yl]oxyphenyl]pyrazol-1-yl]propan-1-ol, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2023-09-29 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Amino Alcohols as Highly Potent, Selective, and Orally Efficacious Inhibitors of Leukotriene A4 Hydrolase.
J.Med.Chem., 66, 2023
|
|
1COT
 
 | |
2WSL
 
 | | Aged Form of Human Butyrylcholinesterase Inhibited by Tabun Analogue TA4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Carletti, E, Aurbek, N, Gillon, E, Loiodice, M, Nicolet, Y, Fontecilla, J, Masson, P, Thiermann, H, Nachon, F, Worek, F. | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-activity analysis of aging and reactivation of human butyrylcholinesterase inhibited by analogues of tabun.
Biochem. J., 421, 2009
|
|
7YUY
 
 | | Structure of a mutated membrane-bound glycosyltransferase | | Descriptor: | (11R,14S)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, X.L, Yang, P, Zhang, M, Liu, X.T, Yu, H.J. | | Deposit date: | 2022-08-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and mechanistic insights into fungal beta-1,3-glucan synthase FKS1.
Nature, 616, 2023
|
|
7Z8D
 
 | | Structure of Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV by fixed-target serial synchrotron crystallography (room temperature, 26keV) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, ... | | Authors: | Gabdulkhakov, A.G, Selikhanov, G.K, Guenther, S, Petrova, T, Meents, A, Fufina, T.Y, Vasilieva, L.G. | | Deposit date: | 2022-03-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV by fixed-target serial synchrotron crystallography (room temperature, 26keV)
To Be Published
|
|
2WVP
 
 | | Synthetically modified OmpG | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, N-[2-({[5-(DIMETHYLAMINO)NAPHTHALEN-1-YL]SULFONYL}AMINO)ETHYL]-2-IODOACETAMIDE, ... | | Authors: | Grosse, W, Reiss, P, Reitz, S, Cebi, M, Luebben, W, Koert, U, Essen, L.-O. | | Deposit date: | 2009-10-19 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Characterization of a Synthetically Modified Ompg.
Bioorg.Med.Chem., 18, 2010
|
|
