7AE9
 
 | | Crystal structure of mono-AMPylated HEPN(R46E) toxin in complex with MNT antitoxin | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-MONOPHOSPHATE, HEPN toxin, MNT ANTITOXIN | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
6QZR
 
 | | 14-3-3 sigma in complex with FOXO1 pT24 peptide | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Forkhead box protein O1, ... | | Authors: | Ottmann, C, Wolter, M, Lau, R.A. | | Deposit date: | 2019-03-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AMPK and AKT protein kinases hierarchically phosphorylate the N-terminus of the FOXO1 transcription factor, modulating interactions with 14-3-3 proteins.
J.Biol.Chem., 294, 2019
|
|
8WNJ
 
 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Ile-AMP | | Descriptor: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
3S22
 
 | | AMP-C BETA-LACTAMASE (PSEUDOMONAS AERUGINOSA) in complex with an inhibitor | | Descriptor: | Beta-lactamase, CHLORIDE ION, [(2S,3R)-2-formyl-1-{[4-(methylamino)butyl]carbamoyl}pyrrolidin-3-yl]sulfamic acid | | Authors: | Scapin, G, Lu, J, Fitzgerald, P.M.D, Sharma, N. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Side chain SAR of bicyclic Beta-lactamase inhibitors (BLIs). 2. N-Alkylated and open chain analogs of MK-8712
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6NAB
 
 | |
1NJF
 
 | | Nucleotide bound form of an isolated E. coli clamp loader gamma subunit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase III subunit gamma, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Podobnik, M, Weitze, T.F, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2002-12-30 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nucleotide-Induced Conformational Changes in an Isolated Escherichia coli DNA Polymerase III Clamp Loader Subunit
Structure, 11, 2003
|
|
1NJG
 
 | | Nucleotide-free form of an Isolated E. coli Clamp Loader Gamma Subunit | | Descriptor: | DNA polymerase III subunit gamma, SULFATE ION, ZINC ION | | Authors: | Podobnik, M, Weitze, T.F, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2002-12-30 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nucleotide-Induced Conformational Changes in an Isolated Escherichia coli DNA Polymerase III Clamp Loader Subunit
Structure, 11, 2003
|
|
6XB6
 
 | |
6NFE
 
 | |
3I59
 
 | | Crystal structure of MtbCRP in complex with N6-cAMP | | Descriptor: | (2R)-N6-(1-Methyl-2-phenylethyl)adenosine-3',5'-cyclic monophosphate, (2S)-N6-(1-Methyl-2-phenylethyl)adenosine-3',5'-cyclic monophosphate, CHLORIDE ION, ... | | Authors: | Reddy, M.C, Palaninathan, S.K, Bruning, J.B, Thurman, C, Smith, D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-07-03 | | Release date: | 2009-09-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Insights into the Mechanism of the Allosteric Transitions of Mycobacterium tuberculosis cAMP Receptor Protein.
J.Biol.Chem., 284, 2009
|
|
3I62
 
 | |
3I61
 
 | |
4U2Q
 
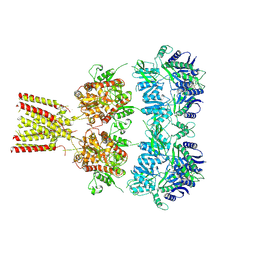 | | Full-length AMPA subtype ionotropic glutamate receptor GluA2 in complex with partial agonist kainate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | Authors: | Duerr, K.L, Chen, L, Gouaux, E. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5247 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
2QCS
 
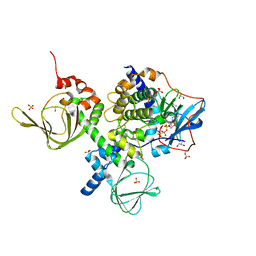 | | A complex structure between the Catalytic and Regulatory subunit of Protein Kinase A that represents the inhibited state | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Kim, C, Cheng, C.Y, Saldanha, A.S, Taylor, S.S. | | Deposit date: | 2007-06-19 | | Release date: | 2007-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PKA-I holoenzyme structure reveals a mechanism for cAMP-dependent activation.
Cell(Cambridge,Mass.), 130, 2007
|
|
4BZP
 
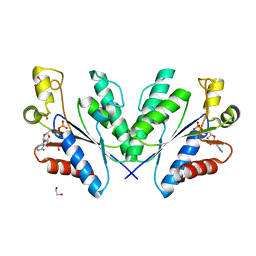 | | Structure of the Mycobacterium tuberculosis APS kinase CysC in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, BIFUNCTIONAL ENZYME CYSN/CYSC | | Authors: | Poyraz, O, Lohkamp, B, Schnell, R, Schneider, G. | | Deposit date: | 2013-07-29 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal Structures of the Kinase Domain of the Sulfate-Activating Complex in Mycobacterium Tuberculosis.
Plos One, 10, 2015
|
|
4U4C
 
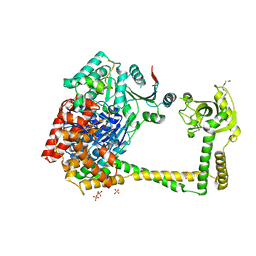 | | The molecular architecture of the TRAMP complex reveals the organization and interplay of its two catalytic activities | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent RNA helicase DOB1, CHLORIDE ION, ... | | Authors: | Falk, S, Weir, J.R, Hentschel, J, Reichelt, P, Bonneau, F, Conti, E. | | Deposit date: | 2014-07-23 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Molecular Architecture of the TRAMP Complex Reveals the Organization and Interplay of Its Two Catalytic Activities.
Mol.Cell, 55, 2014
|
|
4O22
 
 | | Binary complex of metal-free PKAc with SP20. | | Descriptor: | Phosphorylated peptide pSP20., cAMP-dependent protein kinase catalytic subunit alpha. | | Authors: | Das, A, Kovalevsky, A.Y, Gerlits, O, Langan, P, Heller, W.T, Keshwani, M, Taylor, S.S. | | Deposit date: | 2013-12-16 | | Release date: | 2014-05-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal-Free cAMP-Dependent Protein Kinase Can Catalyze Phosphoryl Transfer.
Biochemistry, 53, 2014
|
|
4O21
 
 | | Product complex of metal-free PKAc, ATP-gamma-S and SP20. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thio-phosphorylated peptide pSP20, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Das, A, Kovalevsky, A.Y, Gerlits, O, Langan, P, Heller, W.T, Keshwani, M, Taylor, S. | | Deposit date: | 2013-12-16 | | Release date: | 2014-05-28 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Metal-Free cAMP-Dependent Protein Kinase Can Catalyze Phosphoryl Transfer.
Biochemistry, 53, 2014
|
|
4LV0
 
 | |
9DJ8
 
 | |
3SG7
 
 | | Crystal Structure of GCaMP3-KF(linker 1) | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Schreiter, E.R, Akerboom, J, Looger, L.L. | | Deposit date: | 2011-06-14 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of a GCaMP calcium indicator for neural activity imaging.
J.Neurosci., 32, 2012
|
|
3SG5
 
 | | Crystal Structure of Dimeric GCaMP3-D380Y, QP(linker 1), LP(linker 2) | | Descriptor: | CALCIUM ION, GLYCEROL, Myosin light chain kinase, ... | | Authors: | Schreiter, E.R, Akerboom, J, Looger, L.L. | | Deposit date: | 2011-06-14 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of a GCaMP calcium indicator for neural activity imaging.
J.Neurosci., 32, 2012
|
|
5XPU
 
 | |
3SG2
 
 | | Crystal Structure of GCaMP2-T116V,D381Y | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Schreiter, E.R, Akerboom, J, Looger, L.L. | | Deposit date: | 2011-06-14 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of a GCaMP calcium indicator for neural activity imaging.
J.Neurosci., 32, 2012
|
|
3SG6
 
 | | Crystal Structure of Dimeric GCaMP2-LIA(linker 1) | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Schreiter, E.R, Akerboom, J, Looger, L.L. | | Deposit date: | 2011-06-14 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimization of a GCaMP calcium indicator for neural activity imaging.
J.Neurosci., 32, 2012
|
|
