3CAV
 
 | | Crystal structure of 5beta-reductase (AKR1D1) in complex with NADP+ and 5beta-pregnan-3,20-dione | | Descriptor: | (5BETA)-PREGNANE-3,20-DIONE, 1,2-ETHANEDIOL, 3-oxo-5-beta-steroid 4-dehydrogenase, ... | | Authors: | Faucher, F, Cantin, L, Breton, R. | | Deposit date: | 2008-02-20 | | Release date: | 2008-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of human Delta4-3-ketosteroid 5beta-reductase defines the functional role of the residues of the catalytic tetrad in the steroid double bond reduction mechanism.
Biochemistry, 47, 2008
|
|
3CBW
 
 | | Crystal structure of the YdhT protein from Bacillus subtilis | | Descriptor: | CITRIC ACID, YdhT protein | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-23 | | Release date: | 2008-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.269 Å) | | Cite: | Crystal structure of the YdhT protein from Bacillus subtilis.
To be Published
|
|
2QWC
 
 | |
3CI2
 
 | |
2QWB
 
 | |
3CHQ
 
 | | Crystal structure of leukotriene a4 hydrolase in complex with N5-[4-(phenylmethoxy)phenyl]-L-glutamine | | Descriptor: | (2S)-2-amino-5-oxo-5-[(4-phenylmethoxyphenyl)amino]pentanoic acid, IMIDAZOLE, Leukotriene A-4 hydrolase, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
3CHE
 
 | |
3CI5
 
 | |
3CFJ
 
 | |
2R2L
 
 | | Structure of Farnesyl Protein Transferase bound to PB-93 | | Descriptor: | FARNESYL DIPHOSPHATE, Farnesyltransferase subunit alpha, Farnesyltransferase subunit beta, ... | | Authors: | Strickland, C.O, Voorhis, W. | | Deposit date: | 2007-08-27 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Efficacy, pharmacokinetics, and metabolism of tetrahydroquinoline inhibitors of Plasmodium falciparum protein farnesyltransferase.
Antimicrob.Agents Chemother., 51, 2007
|
|
3CH5
 
 | | The crystal structure of the RanGDP-Nup153ZnF2 complex | | Descriptor: | Fragment of Nuclear pore complex protein Nup153, GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vetter, I.R, Schrader, N. | | Deposit date: | 2008-03-07 | | Release date: | 2008-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Ran-Nup153ZnF2 Complex: a General Ran Docking Site at the Nuclear Pore Complex
Structure, 16, 2008
|
|
3CHR
 
 | | Crystal structure of leukotriene A4 hydrolase in complex with 4-amino-N-[4-(phenylmethoxy)phenyl]-butanamide | | Descriptor: | 4-amino-N-[4-(benzyloxy)phenyl]butanamide, IMIDAZOLE, Leukotriene A-4 hydrolase, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
2QI9
 
 | | ABC-transporter BtuCD in complex with its periplasmic binding protein BtuF | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, PHOSPHATE ION, ... | | Authors: | Hvorup, R.N, Goetz, B.A, Niederer, M, Hollenstein, K, Perozo, E, Locher, K.P. | | Deposit date: | 2007-07-03 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Asymmetry in the structure of the ABC transporter-binding protein complex BtuCD-BtuF.
Science, 317, 2007
|
|
2QJM
 
 | | Crystal structure of the K271E mutant of Mannonate dehydratase from Novosphingobium aromaticivorans complexed with Mg and D-mannonate | | Descriptor: | D-MANNONIC ACID, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Vick, J.E, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-07-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: D-Mannonate dehydratase from Novosphingobium aromaticivorans.
Biochemistry, 46, 2007
|
|
2R0M
 
 | | The effect of a Glu370Asp Mutation in Glutaryl-CoA Dehydrogenase on Proton Transfer to the Dienolate Intermediate | | Descriptor: | 4-nitrobutanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase | | Authors: | Rao, K.S, Fu, Z, Albro, M, Narayanan, B, Baddam, S, Lee, H.J, Kim, J.J, Frerman, F.E. | | Deposit date: | 2007-08-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The effect of a Glu370Asp mutation in glutaryl-CoA dehydrogenase on proton transfer to the dienolate intermediate.
Biochemistry, 46, 2007
|
|
2QQS
 
 | |
2QWD
 
 | |
2QWK
 
 | |
2QB3
 
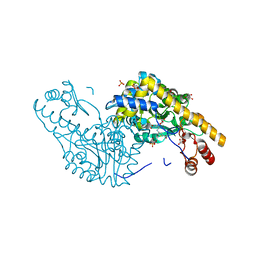 | | Structural Studies Reveal the Inactivation of E. coli L-Aspartate Aminotransferase by (s)-4,5-dihydro-2-thiophenecarboxylic acid (SADTA) via Two Mechanisms (at pH 7.5) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-15 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Inactivation of Escherichia coli L-aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-thiophenecarboxylic acid reveals "a tale of two mechanisms".
Biochemistry, 46, 2007
|
|
2QCB
 
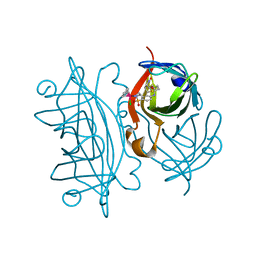 | | T7-tagged full-length streptavidin complexed with ruthenium ligand | | Descriptor: | N-(4-{[(2-AMINOETHYL)AMINO]SULFONYL}PHENYL)-5-[(3AS,4S,6AR)-2-OXOHEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL]PENTANAMIDE-(1,2,3,4,5,6-ETA)-BENZENE-CHLORO-RUTHENIUM(III), Streptavidin | | Authors: | Le Trong, I, Creus, M, Pordea, A, Ward, T.R, Stenkamp, R.E. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structure and designed evolution of an artificial transfer hydrogenase
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
2QJP
 
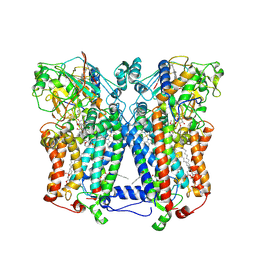 | | Crystal structure of wild type rhodobacter sphaeroides with stigmatellin and antimycin inhibited | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, (2R,3S,6S,7R,8R)-3-{[3-(FORMYLAMINO)-2-HYDROXYBENZOYL]AMINO}-8-HEXYL-2,6-DIMETHYL-4,9-DIOXO-1,5-DIOXONAN-7-YL (2S)-2-METHYLBUTANOATE, 2-O-octyl-beta-D-glucopyranose, ... | | Authors: | Esser, L, Xia, D. | | Deposit date: | 2007-07-08 | | Release date: | 2007-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Inhibitor-complexed structures of the cytochrome bc1 from the photosynthetic bacterium Rhodobacter sphaeroides.
J.Biol.Chem., 283, 2008
|
|
2QLW
 
 | |
2QJJ
 
 | | Crystal structure of D-Mannonate dehydratase from Novosphingobium aromaticivorans | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Vick, J.E, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2007-07-07 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: D-Mannonate dehydratase from Novosphingobium aromaticivorans.
Biochemistry, 46, 2007
|
|
2QWA
 
 | |
2QXF
 
 | |
