6NVU
 
 | | Crystal structure of TLA-1 extended spectrum Beta-lactamase in complex with Clavulanic Acid | | Descriptor: | (2R,3Z,5R)-3-(2-HYDROXYETHYLIDENE)-7-OXO-4-OXA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, 2,5-bis(2-hydroxyethyl)-1,3-oxazole-4-carboxylic acid, ACETATE ION, ... | | Authors: | Rudino-Pinera, E, Cifuentes-Castro, V.H, Rodriguez-Almazan, C. | | Deposit date: | 2019-02-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of ESBL TLA-1 in complex with clavulanic acid reveals a second acylation site.
Biochem.Biophys.Res.Commun., 522, 2020
|
|
8OEG
 
 | | PDE4B bound to MAPI compound 92a | | Descriptor: | MAGNESIUM ION, ZINC ION, [(1~{S})-2-[3,5-bis(chloranyl)-1-oxidanyl-pyridin-4-yl]-1-(3,4-dimethoxyphenyl)ethyl] 5-[[[(1~{R})-2-[[(3~{R})-1-azabicyclo[2.2.2]octan-3-yl]oxy]-2-oxidanylidene-1-phenyl-ethyl]amino]methyl]thiophene-2-carboxylate, ... | | Authors: | Rizzi, A, Armani, E. | | Deposit date: | 2023-03-10 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Optimization of M 3 Antagonist-PDE4 Inhibitor (MAPI) Dual Pharmacology Molecules for the Treatment of COPD.
J.Med.Chem., 66, 2023
|
|
8ODW
 
 | | Crystal structure of LbmA Ox-ACP didomain in complex with NADP and ethyl glycinate from the lobatamide PKS (Gynuella sunshinyii) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Francois, R.M.M, Fraley, A.E, Piel, J, Weissman, K.J, Gruez, A. | | Deposit date: | 2023-03-09 | | Release date: | 2023-05-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Modular Oxime Formation by a trans-AT Polyketide Synthase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
1ON9
 
 | | Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core (with hydrolyzed methylmalonyl-coenzyme a bound) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CADMIUM ION, METHYLMALONYL-COENZYME A, ... | | Authors: | Hall, P.R, Wang, Y.-F, Rivera-Hainaj, R.E, Zheng, X, Pustai-Carey, M, Carey, P.R, Yee, V.C. | | Deposit date: | 2003-02-27 | | Release date: | 2003-05-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Transcarboxylase 12S crystal structure: hexamer assembly and substrate binding to a multienzyme core
Embo J., 22, 2003
|
|
5L5I
 
 | |
2PRJ
 
 | | Binding of N-acetyl-beta-D-glucopyranosylamine to Glycogen Phosphorylase B | | Descriptor: | GLYCOGEN PHOSPHORYLASE, INOSINIC ACID, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Oikonomakos, N.G, Kontou, M, Zographos, S.E, Watson, K.A, Johnson, L.N, Bichard, C.J.F, Fleet, G.W.J, Acharya, K.R. | | Deposit date: | 1998-12-11 | | Release date: | 1998-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N-acetyl-beta-D-glucopyranosylamine: a potent T-state inhibitor of glycogen phosphorylase. A comparison with alpha-D-glucose.
Protein Sci., 4, 1995
|
|
5L60
 
 | |
5L5A
 
 | | Yeast 20S proteasome with human beta5i (1-138; R57T) | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
8W26
 
 | |
5L5U
 
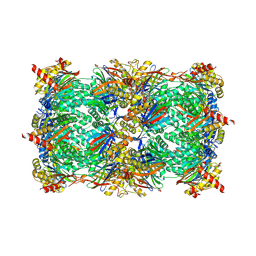 | | Yeast 20S proteasome with human beta5i (1-138; V31M) and human beta6 (97-111; 118-133) in complex with epoxyketone inhibitor 17 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
6NYE
 
 | |
5L69
 
 | | Yeast 20S proteasome with mouse beta5i (1-138) and mouse beta6 (97-111; 118-133) in complex with epoxyketone inhibitor 16 | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-~{N}-[(2~{S},3~{S},4~{R})-4-methyl-3,5-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{R})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
3TAB
 
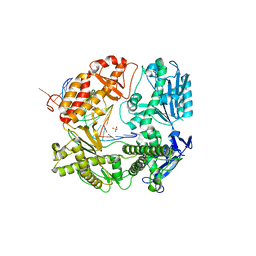 | | 5-hydroxycytosine paired with dGMP in RB69 gp43 | | Descriptor: | DNA (5'-D(*CP*CP*(5OC)P*GP*GP*TP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*AP*CP*CP*G)-3'), DNA polymerase, ... | | Authors: | Zahn, K.E. | | Deposit date: | 2011-08-03 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The miscoding potential of 5-hydroxycytosine arises due to template instability in the replicative polymerase active site.
Biochemistry, 50, 2011
|
|
7M4U
 
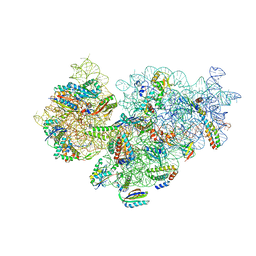 | | A. baumannii Ribosome-Eravacycline complex: 30S | | Descriptor: | 16s Ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
1CEJ
 
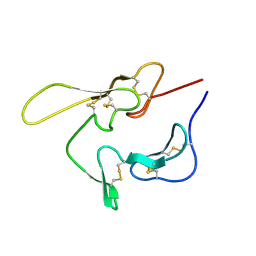 | | SOLUTION STRUCTURE OF AN EGF MODULE PAIR FROM THE PLASMODIUM FALCIPARUM MEROZOITE SURFACE PROTEIN 1 | | Descriptor: | PROTEIN (MEROZOITE SURFACE PROTEIN 1) | | Authors: | Morgan, W.D, Birdsall, B, Frenkiel, T.A, Gradwell, M.G, Burghaus, P.A, Syed, S.E.H, Uthaipibull, C, Holder, A.A, Feeney, J. | | Deposit date: | 1999-03-08 | | Release date: | 1999-05-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an EGF module pair from the Plasmodium falciparum merozoite surface protein 1.
J.Mol.Biol., 289, 1999
|
|
7M4Z
 
 | | A. baumannii Ribosome-Eravacycline complex: hpf-bound 70S | | Descriptor: | 16s Ribosomal RNA, 23s ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
8WT3
 
 | |
2O3O
 
 | |
6O0O
 
 | | crystal structure of BCL-2 G101V mutation with S55746 | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, ~{N}-(4-hydroxyphenyl)-3-[6-[[(3~{S})-3-(morpholin-4-ylmethyl)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]-1,3-benzodioxol-5-yl]-~{N}-phenyl-5,6,7,8-tetrahydroindolizine-1-carboxamide | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
5EJJ
 
 | | Crystal structure of UfSP from C.elegans | | Descriptor: | Ufm1-specific protease | | Authors: | Kim, K, Ha, B, Kim, E.E. | | Deposit date: | 2015-11-02 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The MPN domain of Caenorhabditis elegans UfSP modulates both substrate recognition and deufmylation activity
Biochem. Biophys. Res. Commun., 476, 2016
|
|
3NCF
 
 | | A mutant human Prolactin receptor antagonist H30A in complex with the mutant extracellular domain H188A of the human prolactin receptor | | Descriptor: | CHLORIDE ION, Prolactin, Prolactin receptor, ... | | Authors: | Kulkarni, M.V, Tettamanzi, M.C, Murphy, J.W, Keeler, C, Myszka, D.G, Chayen, N.E, Lolis, E.J, Hodsdon, M.E. | | Deposit date: | 2010-06-04 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two Independent Histidines, One in Human Prolactin and One in Its Receptor, Are Critical for pH-dependent Receptor Recognition and Activation.
J.Biol.Chem., 285, 2010
|
|
8OG4
 
 | | Exostosin-like 3 UDP complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Exostosin-like 3, MANGANESE (II) ION, ... | | Authors: | Sammon, D, Hohenester, E. | | Deposit date: | 2023-03-18 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of decision-making in glycosaminoglycan biosynthesis.
Nat Commun, 14, 2023
|
|
2O7D
 
 | | Tyrosine ammonia-lyase from Rhodobacter sphaeroides, complexed with caffeate | | Descriptor: | CAFFEIC ACID, Putative histidine ammonia-lyase | | Authors: | Louie, G.V, Bowman, M.E, Moffitt, M.C, Baiga, T.J, Moore, B.S, Noel, J.P. | | Deposit date: | 2006-12-10 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases.
Chem.Biol., 13, 2006
|
|
6NZY
 
 | |
3UP3
 
 | | Nuclear receptor DAF-12 from hookworm Ancylostoma ceylanicum in complex with (25S)-cholestenoic acid | | Descriptor: | (8alpha,10alpha,25S)-3-hydroxycholesta-3,5-dien-26-oic acid, 1,2-ETHANEDIOL, Nuclear receptor coactivator 2, ... | | Authors: | Zhi, X, Zhou, X.E, Melcher, K, Motola, D.L, Gelmedin, V, Hawdon, J, Kliewer, S.A, Mangelsdorf, D.J, Xu, H.E. | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Conservation of Ligand Binding Reveals a Bile Acid-like Signaling Pathway in Nematodes.
J.Biol.Chem., 287, 2012
|
|
