6Z6M
 
 | | Cryo-EM structure of human 80S ribosomes bound to EBP1, eEF2 and SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
6Z6N
 
 | | Cryo-EM structure of human EBP1-80S ribosomes (focus on EBP1) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
8XSX
 
 | | Cryo-EM structure of the human 80S ribosome with Tigecycline, E-tRNA, SERBP1 and eEF2 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XSZ
 
 | | Cryo-EM structure of the human 80S ribosome with Tigecycline, E-tRNA and P-tRNA | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XSY
 
 | | Cryo-EM structure of the human 80S ribosome with Tigecycline, e-tRNA and CCDC124 (40S head Swivelled) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8DD7
 
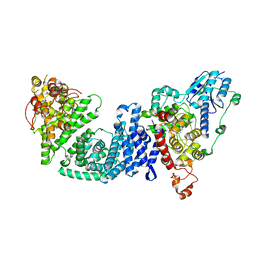 | | The Cryo-EM structure of Drosophila Cryptochrome in complex with Timeless | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methylated-DNA--protein-cysteine methyltransferase,Cryptochrome-1 fusion, Protein timeless,Methylated-DNA--protein-cysteine methyltransferase fusion | | Authors: | Feng, S, Lin, C, DeOliveira, C.C, Crane, B.R. | | Deposit date: | 2022-06-17 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryptochrome-Timeless structure reveals circadian clock timing mechanisms.
Nature, 617, 2023
|
|
5DGE
 
 | | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Melnikov, S, Mailliot, J, Shin, B.-S, Rigger, L, Yusupova, G, Micura, R, Dever, T.E, Yusupov, M. | | Deposit date: | 2015-08-27 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome
To Be Published
|
|
5OBM
 
 | | Crystal structure of Gentamicin bound to the yeast 80S ribosome | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Prokhorova, I, Djumagulov, M, Urzhumtsev, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2017-06-28 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Aminoglycoside interactions and impacts on the eukaryotic ribosome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ON6
 
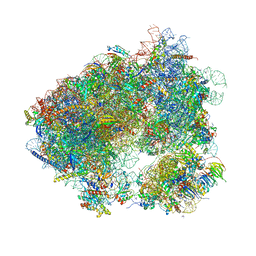 | | Crystal structure of haemanthamine bound to the 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Pellegrino, S, Meyer, M, Yusupova, G, Yusupov, M. | | Deposit date: | 2017-08-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.10000229 Å) | | Cite: | The Amaryllidaceae Alkaloid Haemanthamine Binds the Eukaryotic Ribosome to Repress Cancer Cell Growth.
Structure, 26, 2018
|
|
5DAT
 
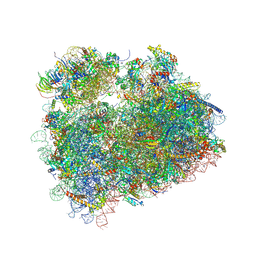 | | Complex of yeast 80S ribosome with hypusine-containing eIF5A | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Melnikov, S, Mailliot, J, Shin, B.-S, Rigger, L, Yusupova, G, Micura, R, Dever, T.E, Yusupov, M. | | Deposit date: | 2015-08-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome
To Be Published
|
|
7K1A
 
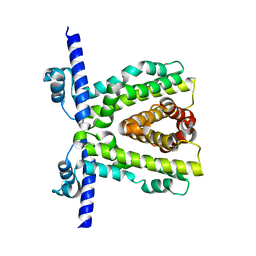 | | TtgR quadruple mutant (C137I I141W M167L F168Y) | | Descriptor: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION | | Authors: | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
7K1C
 
 | | TtgR in complex with resveratrol | | Descriptor: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION, RESVERATROL | | Authors: | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | Deposit date: | 2020-09-07 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
7KD8
 
 | | TtgR C137I I141W M167L F168Y mutant in complex with resveratrol | | Descriptor: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION, RESVERATROL | | Authors: | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | Deposit date: | 2020-10-08 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
8F2I
 
 | | P53 monomer structure | | Descriptor: | Cellular tumor antigen p53 | | Authors: | Solares, M, Kelly, D.F. | | Deposit date: | 2022-11-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | High-Resolution Imaging of Human Cancer Proteins Using Microprocessor Materials.
Chembiochem, 23, 2022
|
|
8F2H
 
 | |
7Z3X
 
 | | Crystal structure of FIR RRM1-2 Y115F mutant bound to FUSE ssDNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*T)-3'), DNA (5'-D(P*GP*TP*())-3'), ... | | Authors: | Ni, X, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-02 | | Release date: | 2022-11-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of FIR RRM1-2 Y115F mutant bound to FUSE ssDNA
To Be Published
|
|
1K61
 
 | | MATALPHA2 HOMEODOMAIN BOUND TO DNA | | Descriptor: | 5'-D(*(5IU)P*GP*CP*GP*TP*GP*TP*AP*AP*AP*TP*GP*AP*AP*TP*TP*AP*CP*AP*TP*G)-3', 5'-D(*AP*CP*AP*TP*GP*TP*AP*AP*TP*TP*CP*AP*TP*TP*TP*AP*CP*AP*CP*GP*C)-3', Mating-type protein alpha-2 | | Authors: | Aishima, J, Gitti, R.K, Noah, J.E, Gan, H.H, Schlick, T, Wolberger, C. | | Deposit date: | 2001-10-14 | | Release date: | 2002-12-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Hoogsteen base pair embedded in undistorted B-DNA
NUCLEIC ACIDS RES., 30, 2002
|
|
2DB7
 
 | | Crystal structure of hypothetical protein MS0332 | | Descriptor: | Hairy/enhancer-of-split related with YRPW motif 1 | | Authors: | Wang, H, Takemoto-Hori, C, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of hypothetical protein MS0332
To be Published
|
|
3NI3
 
 | | 54-Membered ring macrocyclic beta-sheet peptide | | Descriptor: | 54-membered ring macrocyclic beta-sheet peptide, ISOPROPYL ALCOHOL | | Authors: | Sawaya, M.R, Eisenberg, D, Nowick, J.S, Korman, T.P, Khakshoor, O. | | Deposit date: | 2010-06-15 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | X-ray crystallographic structure of an artificial beta-sheet dimer.
J.Am.Chem.Soc., 132, 2010
|
|
3ABU
 
 | | Crystal Structure of LSD1 in complex with a 2-PCPA derivative, S1201 | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-[(1R,3R,3aS)-3-[2-(benzyloxy)-3-fluorophenyl]-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate | | Authors: | Mimasu, S, Umezawa, N, Sato, S, Higuchi, T, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-21 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structurally Designed trans-2-Phenylcyclopropylamine Derivatives Potently Inhibit Histone Demethylase LSD1/KDM1
Biochemistry, 49, 2010
|
|
3ABT
 
 | | Crystal Structure of LSD1 in complex with trans-2-pentafluorophenylcyclopropylamine | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(1R,3R,3aS)-1-hydroxy-10,11-dimethyl-4,6-dioxo-3-(pentafluorophenyl)-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]pentyl dihydrogen diphosphate | | Authors: | Mimasu, S, Umezawa, N, Sato, S, Higuchi, T, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-21 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structurally Designed trans-2-Phenylcyclopropylamine Derivatives Potently Inhibit Histone Demethylase LSD1/KDM1
Biochemistry, 49, 2010
|
|
3B5B
 
 | | Crystal structure of the thymidylate synthase k48q | | Descriptor: | 2'-DEOXY-5-NITROURIDINE 5'-(DIHYDROGEN PHOSPHATE), 2'-DEOXY-5-NITROURIDINE 5'-MONOPHOSPHATE, FORMIC ACID, ... | | Authors: | Sotelo-Mundo, R.R, Arreola, R, Maley, F, Montfort, W.R. | | Deposit date: | 2007-10-25 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Role of an invariant lysine residue in folate binding on Escherichia coli thymidylate synthase: Calorimetric and crystallographic analysis of the K48Q mutant.
Int.J.Biochem.Cell Biol., 40, 2008
|
|
4V56
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with spectinomycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Shoji, S, Holton, J.M, Fredrick, K, Cate, J.H.D. | | Deposit date: | 2007-07-21 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.93 Å) | | Cite: | A steric block in translation caused by the antibiotic spectinomycin.
Acs Chem.Biol., 2, 2007
|
|
3NXT
 
 | | Preferential Selection of Isomer Binding from Chiral Mixtures: Alternate Binding Modes Observed for the E-and Z-isomers of a Series of 5-substituted 2,4-diaminofuro[2m,3-d]pyrimidines as Ternary Complexes with NADPH and Human Dihydrofolate Reductase | | Descriptor: | 5-[(E)-2-cyclopropyl-2-(2-methoxyphenyl)ethenyl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2010-07-14 | | Release date: | 2010-12-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Preferential selection of isomer binding from chiral mixtures: alternate binding modes observed for the E and Z isomers of a series of 5-substituted 2,4-diaminofuro[2,3-d]pyrimidines as ternary complexes with NADPH and human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6QXH
 
 | |
