7T65
 
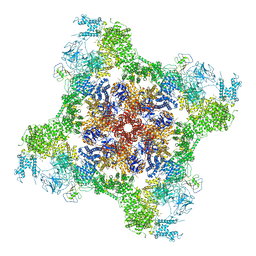 | | Rabbit RyR1 disease mutant Y523S in complex with FKBP12.6 embedded in lipidic nanodisc in the open state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Iyer, K.A, Hu, Y, Murayama, T, Samso, M. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Molecular mechanism of the severe MH/CCD mutation Y522S in skeletal ryanodine receptor (RyR1) by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1NG8
 
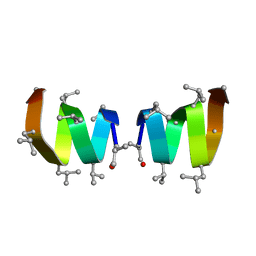 | |
7K0T
 
 | |
1NT6
 
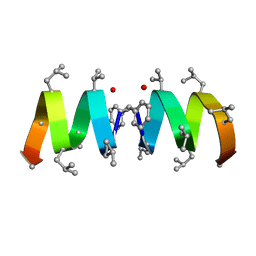 | | F1-Gramicidin C In Sodium Dodecyl Sulfate Micelles (NMR) | | Descriptor: | GRAMICIDIN C | | Authors: | Townsley, L.E, Fletcher, T.G, Hinton, J.F. | | Deposit date: | 2003-01-28 | | Release date: | 2003-02-11 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The Structure, Cation Binding, Transport, and Conductance of Gly15-Gramicidin a Incorporated Into Sds Micelles and Pc/Pg Vesicles.
Biochemistry, 42, 2003
|
|
1NT5
 
 | | F1-Gramicidin A in Sodium Dodecyl Sulfate Micelles (NMR) | | Descriptor: | GRAMICIDIN A | | Authors: | Townsley, L.E, Fletcher, T.G, Hinton, J.F. | | Deposit date: | 2003-01-28 | | Release date: | 2003-02-11 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The Structure, Cation Binding, Transport, and Conductance of Gly15-Gramicidin a Incorporated Into Sds Micelles and Pc/Pg Vesicles.
Biochemistry, 42, 2003
|
|
5TP6
 
 | |
5TP5
 
 | |
6QPB
 
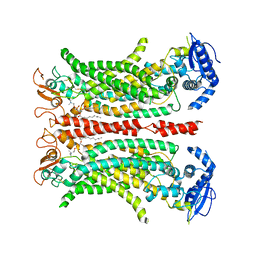 | | Cryo-EM structure of calcium-free mTMEM16F lipid scramblase in digitonin | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6 | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-13 | | Release date: | 2019-03-06 | | Last modified: | 2019-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
6QP6
 
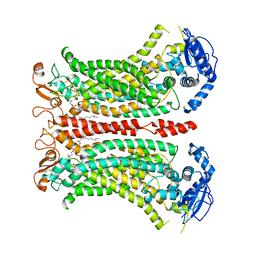 | | Cryo-EM structure of calcium-bound mTMEM16F lipid scramblase in digitonin | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6, CALCIUM ION | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-13 | | Release date: | 2019-03-06 | | Last modified: | 2019-04-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
6QPI
 
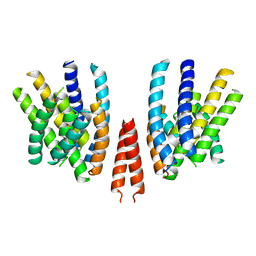 | | Cryo-EM structure of calcium-free mTMEM16F lipid scramblase in nanodisc | | Descriptor: | Anoctamin-6 | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
6QPC
 
 | | Cryo-EM structure of calcium-bound mTMEM16F lipid scramblase in nanodisc | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6, CALCIUM ION | | Authors: | Alvadia, C, Lim, N.K, Clerico Mosina, V, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-13 | | Release date: | 2019-03-06 | | Last modified: | 2019-04-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F.
Elife, 8, 2019
|
|
5A6R
 
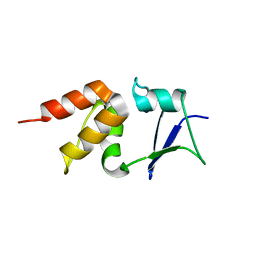 | | Crystal structure of the BTB domain of human KCTD17 | | Descriptor: | BTB/POZ DOMAIN-CONTAINING PROTEIN KCTD17 | | Authors: | Pinkas, D.M, Sorrell, F, Sanvitale, C.E, Goubin, S, Williams, E, Newman, J.A, Pearce, N.M, Neshich, I, Pike, A.C.W, MacKenzie, A, Quigley, A, Faust, B, Carpenter, E.P, Tallant, C, Kopec, J, Chalk, R, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
4CRH
 
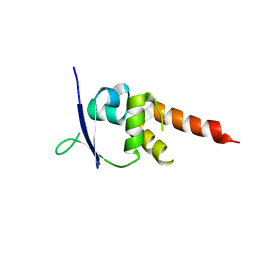 | | Crystal structure of the BTB-T1 domain of human SHKBP1 | | Descriptor: | SH3KBP1-BINDING PROTEIN 1 | | Authors: | Pinkas, D.M, Solcan, N, Krojer, T, Goubin, S, Williams, E.P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
1DTX
 
 | |
5K3S
 
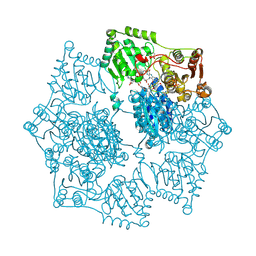 | | Crystal structure of Arabidopsis thaliana acetohydroxyacid synthase in complex with a pyrimidinyl-benzoate herbicide, bispyribac-sodium | | Descriptor: | (3Z)-4-{[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]AMINO}-3-MERCAPTOPENT-3-EN-1-YL TRIHYDROGEN DIPHOSPHATE, 2,6-bis[(4,6-dimethoxypyrimidin-2-yl)oxy]benzoic acid, Acetolactate synthase, ... | | Authors: | Garcia, M.D, Lonhienne, T, Guddat, L.W. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.873 Å) | | Cite: | Comprehensive understanding of acetohydroxyacid synthase inhibition by different herbicide families.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KCV
 
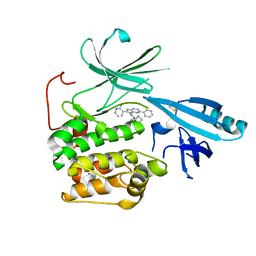 | | Crystal structure of allosteric inhibitor, ARQ 092, in complex with autoinhibited form of AKT1 | | Descriptor: | 3-[3-[4-(1-azanylcyclobutyl)phenyl]-5-phenyl-imidazo[4,5-b]pyridin-2-yl]pyridin-2-amine, RAC-alpha serine/threonine-protein kinase | | Authors: | Eathiraj, S. | | Deposit date: | 2016-06-07 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of 3-(3-(4-(1-Aminocyclobutyl)phenyl)-5-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amine (ARQ 092): An Orally Bioavailable, Selective, and Potent Allosteric AKT Inhibitor.
J.Med.Chem., 59, 2016
|
|
6J95
 
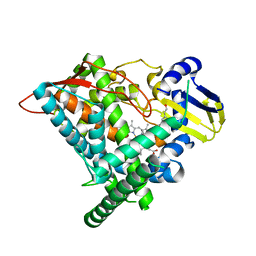 | | Crystal structure of CYP97A3 in complex with retinal | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Protein LUTEIN DEFICIENT 5, chloroplastic, ... | | Authors: | Niu, G, Guo, Q, Wang, J, Zhao, S. | | Deposit date: | 2019-01-22 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structural basis for plant lutein biosynthesis from alpha-carotene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XOG
 
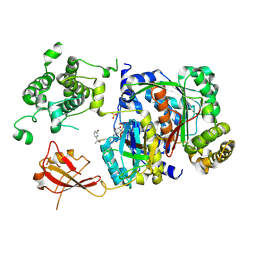 | | Structure of SUMO1-ML786519 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XOH
 
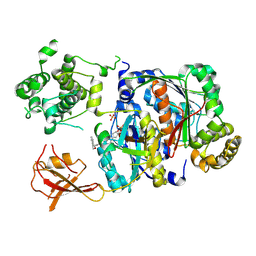 | | Structure of SUMO1-ML00789344 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XOI
 
 | | Structure of SUMO1-ML00752641 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
7ZL3
 
 | |
5J8H
 
 | | Structure of calmodulin in a complex with a peptide derived from a calmodulin-dependent kinase | | Descriptor: | CALCIUM ION, Calmodulin, Eukaryotic elongation factor 2 kinase | | Authors: | Alphonse, S, Lee, K, Piserchio, A, Tavares, C.D.J, Giles, D.H, Wellmann, R.M, Dalby, K.N, Ghose, R. | | Deposit date: | 2016-04-07 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Recognition of Eukaryotic Elongation Factor 2 Kinase by Calmodulin.
Structure, 24, 2016
|
|
5IPO
 
 | |
5JYH
 
 | |
6SGA
 
 | | Body domain of the mt-SSU assemblosome from Trypanosoma brucei | | Descriptor: | 9S rRNA, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Saurer, M, Ramrath, D.J.F, Niemann, M, Calderaro, S, Prange, C, Mattei, S, Scaiola, A, Leitner, A, Bieri, P, Horn, E.K, Leibundgut, M, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2019-08-03 | | Release date: | 2019-09-18 | | Last modified: | 2019-09-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mitoribosomal small subunit biogenesis in trypanosomes involves an extensive assembly machinery.
Science, 365, 2019
|
|
