7Y4N
 
 | | Insight into the C-terminal SH3 domain mediated binding of Drosophila Drk to Sos and Dos | | Descriptor: | Growth factor receptor-bound protein 2 | | Authors: | Pooppadi, M.S, Ikeya, T, Sugasawa, H, Watanabe, R, Mishima, M, Inomata, K, Ito, Y. | | Deposit date: | 2022-06-15 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insight into the C-terminal SH3 domain mediated binding of Drosophila Drk to Sos and Dos.
Biochem.Biophys.Res.Commun., 625, 2022
|
|
3PB5
 
 | | Endothiapepsin in complex with a fragment | | Descriptor: | Endothiapepsin, GLYCEROL, N-methyl-1-[5-(pyridin-3-yloxy)furan-2-yl]methanamine | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-10-20 | | Release date: | 2011-10-19 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A small nonrule of 3 compatible fragment library provides high hit rate of endothiapepsin crystal structures with various fragment chemotypes.
J.Med.Chem., 54, 2011
|
|
7YF1
 
 | |
3OO0
 
 | | Structure of apo CheY A113P | | Descriptor: | AMMONIUM ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Pazy, Y, Collins, E.J, Guanga, G.P, Miller, P.J, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2010-08-30 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Exploring the effect of an allosteric site on conformational coupling in CheY
To be Published
|
|
3OQK
 
 | | Crystal Structure Analysis of Renin-indole-piperazin inhibitor complexes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-phenoxy-1-phenyl-3-(piperazin-1-ylcarbonyl)-1H-indole, GLYCEROL, ... | | Authors: | Bocskei, Z. | | Deposit date: | 2010-09-03 | | Release date: | 2010-10-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and optimization of a new class of potent and non-chiral indole-3-carboxamide-based renin inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3ORD
 
 | |
3OLW
 
 | |
7XQV
 
 | | The complex of nanobody Rh57 binding to GTP-bound RhoA active form | | Descriptor: | ALANINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Zhang, Y.R, Liu, R, Ding, Y. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural insights into the binding of nanobody Rh57 to active RhoA-GTP.
Biochem.Biophys.Res.Commun., 616, 2022
|
|
3OLX
 
 | |
2VJ7
 
 | | Human BACE-1 in complex with 3-(ethylamino)-N-((1S,2R)-2-hydroxy-1-(phenylmethyl)-3-(((3-(trifluoromethyl)phenyl)methyl)amino)propyl)-5-(2-oxo-1-pyrrolidinyl)benzamide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-2-hydroxy-3-{[3-(trifluoromethyl)benzyl]amino}propyl]-3-(ethylamino)-5-(2-oxopyrrolidin-1-yl)benzamide | | Authors: | Clarke, B, Demont, E, Dingwall, C, Dunsdon, R, Faller, A, Hawkins, J, Hussain, I, MacPherson, D, Maile, G, Matico, R, Milner, P, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Riddell, D, Rowland, P, Soleil, V, Smith, K, Stanway, S, Stemp, G, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Ward, J, Wayne, G. | | Deposit date: | 2007-12-06 | | Release date: | 2008-01-29 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bace-1 Inhibitors Part 2: Identification of Hydroxy Ethylamines (Heas) with Reduced Peptidic Character.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7XSV
 
 | | Crystal Structures of PIM1 in Complex with Macrocyclic Compound H3 | | Descriptor: | 8-Methyl-2,5,20-trioxa-8,13,17-triazatetracyclo[11.10.2.014,19.021,25]pentacosa-1(24),14(19),15,17,21(25),22-hexaene, Serine/threonine-protein kinase pim-1 | | Authors: | Shen, C, Xie, Y, Ren, X, Zhou, Y, Niu, H. | | Deposit date: | 2022-05-15 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Design, synthesis, and bioactivity evaluation of macrocyclic benzo[b]pyrido[4,3-e][1,4]oxazine derivatives as novel Pim-1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 72, 2022
|
|
3MWI
 
 | | The complex crystal Structure of Urokianse and 5-nitro-1H-indole-2-amidine | | Descriptor: | 5-nitro-1H-indole-2-carboximidamide, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Yu, H.Y, Yuan, C, Huang, Z.X, Huang, M.D. | | Deposit date: | 2010-05-06 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The complex crystal Structure of Urokianse and 5-nitro-1H-indole-2-amidine
To be Published
|
|
3MTY
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region A of the crystal. First step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MUR
 
 | | Crystal Structure of the C92U mutant c-di-GMP riboswith bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), C92U mutant c-di-GMP riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
3MUM
 
 | | Crystal Structure of the G20A mutant c-di-GMP riboswith bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), G20A mutant c-di-GMP Riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
2VM0
 
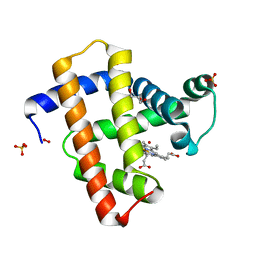 | | Crystal structure of radiation-induced myoglobin compound II generated after annealing of peroxymyoglobin | | Descriptor: | GLYCEROL, HYDROGEN PEROXIDE, HYDROXIDE ION, ... | | Authors: | Hersleth, H.-P, Gorbitz, C.H, Andersson, K.K. | | Deposit date: | 2008-01-20 | | Release date: | 2008-01-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Peroxymyoglobin Generated Through Cryoradiolytic Reduction of Myoglobin Compound III During Data Collection.
Biochem.J., 412, 2008
|
|
3MVS
 
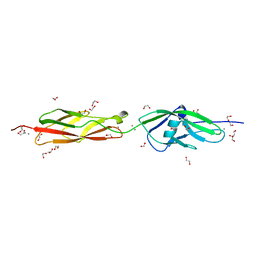 | | Structure of the N-terminus of Cadherin 23 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cadherin-23 | | Authors: | Clark, P, Joseph, J.S, Kolatkar, A.R. | | Deposit date: | 2010-05-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of the N terminus of cadherin 23 reveals a new adhesion mechanism for a subset of cadherin superfamily members.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MZQ
 
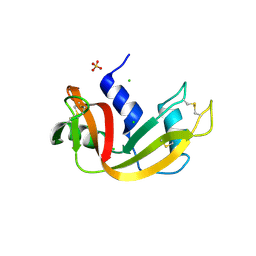 | | RNase crystals grown by the hanging drop method | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Mathews, I.I. | | Deposit date: | 2010-05-12 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diffraction study of protein crystals grown in cryoloops and micromounts.
J.Appl.Crystallogr., 43, 2010
|
|
3MWR
 
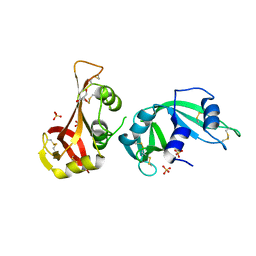 | |
2VIN
 
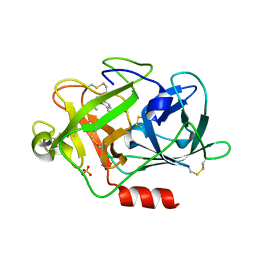 | | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator | | Descriptor: | (2R)-1-(2,6-dimethylphenoxy)propan-2-amine, ACETATE ION, SULFATE ION, ... | | Authors: | Frederickson, M, Callaghan, O, Chessari, G, Congreve, M, Cowan, S.R, Matthews, J.E, McMenamin, R, Smith, D, Vinkovic, M, Wallis, N.G. | | Deposit date: | 2007-12-05 | | Release date: | 2008-01-22 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-Based Discovery of Mexiletine Derivatives as Orally Bioavailable Inhibitors of Urokinase-Type Plasminogen Activator
J.Med.Chem., 51, 2008
|
|
2V0A
 
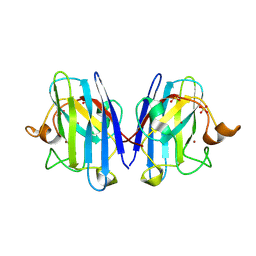 | | Atomic resolution crystal structure of Human Superoxide Dismutase | | Descriptor: | ACETATE ION, COPPER (II) ION, SULFATE ION, ... | | Authors: | Strange, R.W, Antonyuk, S, Yong, C.W, Smith, W, Hasnain, S.S. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Molecular Dynamics Using Atomic-Resolution Structure Reveal Structural Fluctuations that May Lead to Polymerization of Human Cu-Zn Superoxide Dismutase.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
7Y1G
 
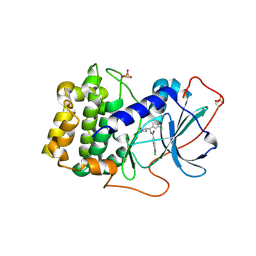 | | Crystal structure of human PRKACA complexed with DS01080522 | | Descriptor: | 1-chloranyl-~{N}-[(~{S})-(3-chloranyl-4-cyano-phenyl)-[(2~{R},4~{S})-4-oxidanylpyrrolidin-2-yl]methyl]-7-methoxy-isoquinoline-6-carboxamide, ZINC ION, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Suzuki, M, Ubukata, O, Toyoda, A. | | Deposit date: | 2022-06-08 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel protein kinase cAMP-Activated Catalytic Subunit Alpha (PRKACA) inhibitor shows anti-tumor activity in a fibrolamellar hepatocellular carcinoma model.
Biochem.Biophys.Res.Commun., 621, 2022
|
|
3MZR
 
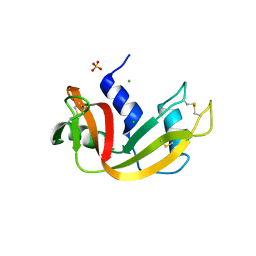 | | RNase crystals grown in loops/micromounts | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Mathews, I.I. | | Deposit date: | 2010-05-12 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diffraction study of protein crystals grown in cryoloops and micromounts.
J.Appl.Crystallogr., 43, 2010
|
|
7Y6M
 
 | |
3N53
 
 | |
