6JWU
 
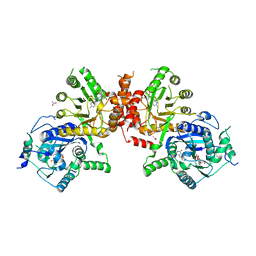 | | Crystal structure of Plasmodium falciparum HPPK-DHPS wild type with STZ-DHP | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 4-{[(2-amino-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)methyl]amino}-N-(1,3-thiazol-2-yl)benzenesulfonamide, 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase-dihydropteroate synthase, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Yuthavong, Y. | | Deposit date: | 2019-04-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The structure of Plasmodium falciparum hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate synthase reveals the basis of sulfa resistance.
Febs J., 287, 2020
|
|
5VOS
 
 | | VGSNKGAIIGL from Amyloid Beta determined by MicroED | | Descriptor: | Amyloid beta A4 protein | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S, Griner, S.L, Gonen, T. | | Deposit date: | 2017-05-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.42 Å) | | Cite: | Common fibrillar spines of amyloid-beta and human islet amyloid polypeptide revealed by microelectron diffraction and structure-based inhibitors.
J. Biol. Chem., 293, 2018
|
|
5ODL
 
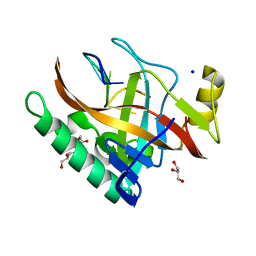 | | Single-stranded DNA-binding protein from bacteriophage Enc34 in complex with ssDNA | | Descriptor: | GLYCEROL, SODIUM ION, oligo(T), ... | | Authors: | Cernooka, E, Rumnieks, J, Kazaks, A, Tars, K. | | Deposit date: | 2017-07-05 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Basis for DNA Recognition of a Single-stranded DNA-binding Protein from Enterobacter Phage Enc34.
Sci Rep, 7, 2017
|
|
5L23
 
 | |
5NTG
 
 | |
5L62
 
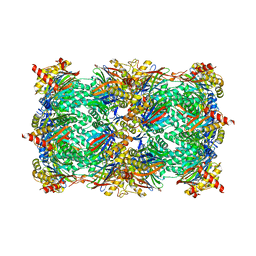 | | Yeast 20S proteasome with human beta5c (1-138) and human beta6 (97-111; 118-133) in complex with epoxyketone inhibitor 16 | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-~{N}-[(2~{S},3~{S},4~{R})-4-methyl-3,5-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{R})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
5VJ9
 
 | |
2WBF
 
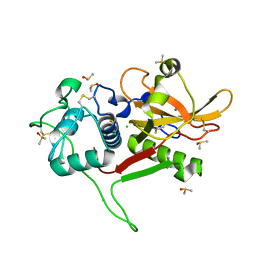 | | Crystal Structure Analysis of SERA5E from plasmodium falciparum with loop 690-700 ordered | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Smith, B.J, Malby, R.L, Colman, P.M, Clarke, O.B. | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the protease-like antigen Plasmodium falciparum SERA5 and its noncanonical active-site serine.
J. Mol. Biol., 392, 2009
|
|
5O5U
 
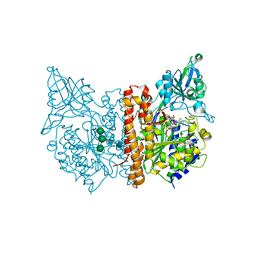 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a urea based inhibitor PSMA 1027 | | Descriptor: | (2~{S})-2-[[(2~{S})-6-[[(2~{S})-2-[[4-[[[(2~{S})-2-[[(2~{R})-2-[(6-fluoranylpyridin-3-yl)carbonylamino]-3-sulfo-propanoyl]amino]-3-sulfo-propanoyl]amino]methyl]phenyl]carbonylamino]-3-naphthalen-2-yl-propanoyl]amino]-1-oxidanyl-1-oxidanylidene-hexan-2-yl]carbamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Novakova, Z, Barinka, C, Motlova, L. | | Deposit date: | 2017-06-02 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a urea based inhibitor PSMA 1027
To Be Published
|
|
5VR6
 
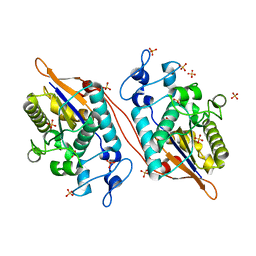 | | Structure of Human Sts-1 histidine phosphatase domain with sulfate bound | | Descriptor: | SULFATE ION, Ubiquitin-associated and SH3 domain-containing protein B | | Authors: | Zhou, W, Yin, Y, Weinheimer, A.W, Kaur, N, Carpino, N, French, J.B. | | Deposit date: | 2017-05-10 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Functional Characterization of the Histidine Phosphatase Domains of Human Sts-1 and Sts-2.
Biochemistry, 56, 2017
|
|
5O6A
 
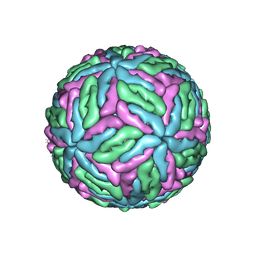 | |
5VAJ
 
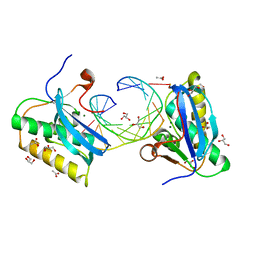 | | BhRNase H - amide-RNA/DNA complex | | Descriptor: | ACETATE ION, DNA (5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3'), GLYCEROL, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-03-27 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Amide linkages mimic phosphates in RNA interactions with proteins and are well tolerated in the guide strand of short interfering RNAs.
Nucleic Acids Res., 45, 2017
|
|
5NY1
 
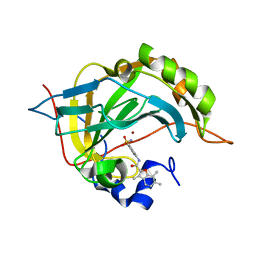 | | Carbonic Anhydrase II Inhibitor RA10 | | Descriptor: | 3,6,7-trimethyl-~{N}-[(4-sulfamoylphenyl)methyl]-1-benzofuran-2-carboxamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5L9K
 
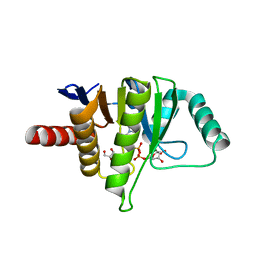 | | OCEANOBACILLUS IHEYENSIS MACRODOMAIN WITH ADPR | | Descriptor: | GLYCEROL, MACROD-TYPE MACRODOMAIN, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Gil-Ortiz, F, Zapata-Perez, R, Martinez, A.B, Juanhuix, J, Sanchez-Ferrer, A. | | Deposit date: | 2016-06-10 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Open Biol, 7, 2017
|
|
1FVF
 
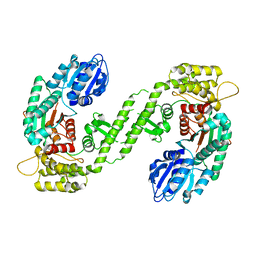 | |
5L9Z
 
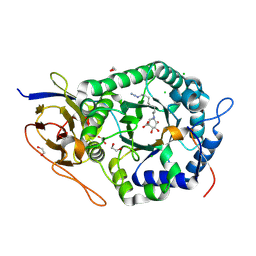 | | Crystal structure of human heparanase nucleophile mutant (E343Q), in complex with unreacted glucuronic acid configured aziridine probe JJB355 | | Descriptor: | (1~{R},2~{S},3~{R},4~{S},5~{S},6~{R})-7-[8-[(azanylidene-{4}-azanylidene)amino]octyl]-3,4,5-tris(oxidanyl)-7-azabicyclo[4.1.0]heptane-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Jin, Y, Davies, G.J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
5NZ4
 
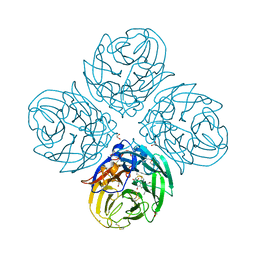 | | Complex of I223V mutant variant of neuraminidase from H1N1 influenza virus with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J, Karlukova, E. | | Deposit date: | 2017-05-12 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Kinetic, Thermodynamic, and Structural Analysis of Drug Resistance Mutations in Neuraminidase from the 2009 Pandemic Influenza Virus.
Viruses, 10, 2018
|
|
5NZW
 
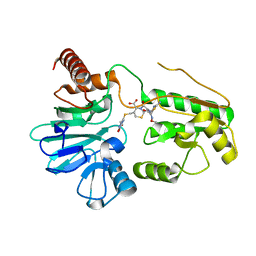 | | Crystal structure of DNA cross-link repair protein 1A in complex with ceftriaxone | | Descriptor: | Ceftriaxone, DNA cross-link repair 1A protein, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Kupinska, K, Burgess-Brown, N.A, Talon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of DNA cross-link repair protein 1A in complex with ceftriaxone
To Be Published
|
|
5UJK
 
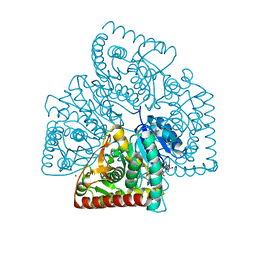 | |
5L5Y
 
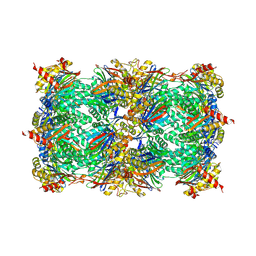 | |
1FZR
 
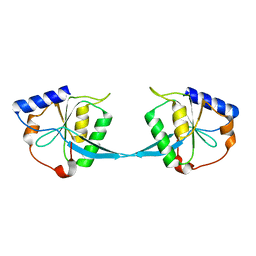 | | CRYSTAL STRUCTURE OF BACTERIOPHAGE T7 ENDONUCLEASE I | | Descriptor: | ENDONUCLEASE I | | Authors: | Hadden, J.M, Convery, M.A, Declais, A.C, Lilley, D.M.J, Phillips, S.E.V. | | Deposit date: | 2000-10-04 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Holliday junction resolving enzyme T7 endonuclease I.
Nat.Struct.Biol., 8, 2001
|
|
6JVV
 
 | |
5UW0
 
 | |
5NYR
 
 | | Anbu from Hyphomicrobium sp. strain MC1 -SG: R3 | | Descriptor: | Anbu, SULFATE ION | | Authors: | Vielberg, M.-T, Groll, M. | | Deposit date: | 2017-05-11 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | On the Trails of the Proteasome Fold: Structural and Functional Analysis of the Ancestral beta-Subunit Protein Anbu.
J. Mol. Biol., 430, 2018
|
|
5L3O
 
 | | Crystal Structure of Human Carbonic Anhydrase II in Complex with a Quinoline Oligoamide Foldamer | | Descriptor: | Aromatic foldamer, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Jewginski, M, Langlois d'Estaintot, B, Granier, T, Huc, Y. | | Deposit date: | 2016-05-24 | | Release date: | 2017-03-01 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Self-Assembled Protein-Aromatic Foldamer Complexes with 2:3 and 2:2:1 Stoichiometries.
J. Am. Chem. Soc., 139, 2017
|
|
