7JOO
 
 | | Crystal structure of ICOS in complex with antibody STIM003 and anti-kappa VHH domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Inducible T-cell costimulator, ... | | Authors: | Rujas, E, Sicard, T, Julien, J.P. | | Deposit date: | 2020-08-06 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of the ICOS/ICOS-L immune complex reveals high molecular mimicry by therapeutic antibodies.
Nat Commun, 11, 2020
|
|
7UMG
 
 | | Crystal structure of human CD8aa-MR1-Ac-6-FP complex | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, Major histocompatibility complex class I-related gene protein, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2022-04-06 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CD8 coreceptor engagement of MR1 enhances antigen responsiveness by human MAIT and other MR1-reactive T cells.
J.Exp.Med., 219, 2022
|
|
5LXW
 
 | |
7V11
 
 | | Factor XIa in Complex with Compound 2e | | Descriptor: | 5-[1-[(1~{R})-1-[5-[3-chloranyl-2-fluoranyl-6-(1,2,3,4-tetrazol-1-yl)phenyl]-1-oxidanyl-pyridin-2-yl]-2-cyclopropyl-ethyl]pyrazol-4-yl]-6-methyl-pyridin-2-amine, CITRIC ACID, Coagulation factor XIa light chain | | Authors: | Shaffer, P.L, Milligan, C.M. | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.472 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Pyridine N-Oxide-Based Factor XIa Inhibitors through Exploiting Nonclassical Interactions.
J.Med.Chem., 65, 2022
|
|
7JWU
 
 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | Descriptor: | 1-methyl-5-phenyl-6-{[(1R)-1-(pyridin-2-yl)ethyl]sulfanyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hurley, T.D, Buchman, C. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
7JPQ
 
 | | ORC-O2-5: Human Origin Recognition Complex (ORC) with subunits 2,3,4,5 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Origin recognition complex subunit 2, ... | | Authors: | Jaremko, M.J, Joshua-Tor, L. | | Deposit date: | 2020-08-09 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The dynamic nature of the human Origin Recognition Complex revealed through five cryoEM structures.
Elife, 9, 2020
|
|
5LVX
 
 | | Crystal structure of glucocerebrosidase with an inhibitory quinazoline modulator | | Descriptor: | 11-[(2~{R})-2-[(2-pyridin-3-ylquinazolin-4-yl)amino]-2,3-dihydro-1~{H}-inden-5-yl]undec-10-ynoic acid, 11-[(2~{S})-2-[(2-pyridin-3-ylquinazolin-4-yl)amino]-2,3-dihydro-1~{H}-inden-5-yl]undec-10-ynoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, J, Chen, L, Skinner, O.S, Lansbury, P, Skerlj, R, Mrosek, M, Heunisch, U, Krapp, S, Weigand, S, Charrow, J, Schwake, M, Kelleher, N.L, Silverman, R.B, Krainc, D. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | beta-Glucocerebrosidase Modulators Promote Dimerization of beta-Glucocerebrosidase and Reveal an Allosteric Binding Site.
J. Am. Chem. Soc., 140, 2018
|
|
7V15
 
 | | Factor XIa in Complex with Compound 2i | | Descriptor: | 5-[1-[(1~{R})-1-[5-[3-chloranyl-2-fluoranyl-6-(1,2,3,4-tetrazol-1-yl)phenyl]-1-oxidanyl-pyridin-2-yl]-2-cyclopropyl-ethyl]pyrazol-4-yl]-4-methyl-1,3-thiazole, CITRIC ACID, Coagulation factor XIa light chain | | Authors: | Shaffer, P.L, Cedervall, P, Milligan, C.M. | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Pyridine N-Oxide-Based Factor XIa Inhibitors through Exploiting Nonclassical Interactions.
J.Med.Chem., 65, 2022
|
|
5M12
 
 | | Structure of GH36 alpha-galactosidase from Thermotoga maritima in complex with intact cyclopropyl-carbasugar. | | Descriptor: | (1~{R},2~{S},3~{S},4~{R},5~{S},6~{S})-5-[3,5-bis(fluoranyl)phenoxy]-1-(hydroxymethyl)bicyclo[4.1.0]heptane-2,3,4-triol, Alpha-galactosidase, MAGNESIUM ION, ... | | Authors: | Pengelly, R, Gloster, T. | | Deposit date: | 2016-10-07 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural Snapshots for Mechanism-Based Inactivation of a Glycoside Hydrolase by Cyclopropyl Carbasugars.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5M0D
 
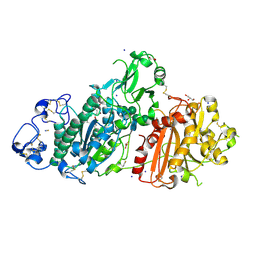 | | Structure-based evolution of a hybrid steroid series of Autotaxin inhibitors | | Descriptor: | CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2,Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, GLYCEROL, ... | | Authors: | Keune, W.-J, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2016-10-04 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational Design of Autotaxin Inhibitors by Structural Evolution of Endogenous Modulators.
J. Med. Chem., 60, 2017
|
|
7KC0
 
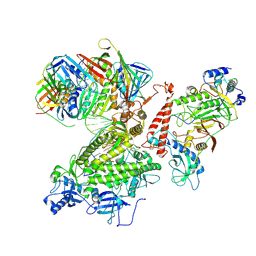 | | Structure of the Saccharomyces cerevisiae replicative polymerase delta in complex with a primer/template and the PCNA clamp | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA (25-MER), DNA (5'-D(P*AP*TP*GP*AP*CP*CP*AP*TP*GP*AP*TP*TP*AP*CP*GP*AP*AP*TP*TP*GP*C)-3'), ... | | Authors: | Zheng, F, Georgescu, R, Li, H, O'Donnell, M.E. | | Deposit date: | 2020-10-04 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of eukaryotic DNA polymerase delta bound to the PCNA clamp while encircling DNA.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5M0O
 
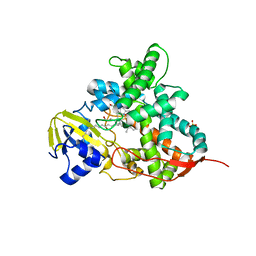 | | Crystal structure of cytochrome P450 OleT H85Q in complex with arachidonic acid | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Tee, K.L, Munro, A, Matthews, S, Leys, D, Levy, C. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic Determinants of Alkene Production by the Cytochrome P450 Peroxygenase OleTJE.
J. Biol. Chem., 292, 2017
|
|
7V10
 
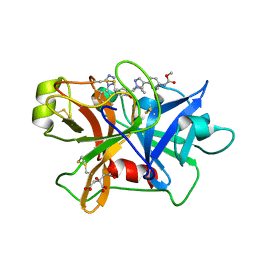 | | Factor XIa in Complex with Compound 2d | | Descriptor: | CITRIC ACID, Coagulation factor XIa light chain, methyl ~{N}-[4-[1-[(1~{R})-1-[5-[3-chloranyl-2-fluoranyl-6-(1,2,3,4-tetrazol-1-yl)phenyl]-1-oxidanyl-pyridin-2-yl]-2-cyclopropyl-ethyl]pyrazol-4-yl]phenyl]carbamate | | Authors: | Shaffer, P.L, Spurlino, J, Milligan, C.M. | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Pyridine N-Oxide-Based Factor XIa Inhibitors through Exploiting Nonclassical Interactions.
J.Med.Chem., 65, 2022
|
|
7UFS
 
 | | Cryo-EM Structure of Bl_Man38B at 3.4 A | | Descriptor: | Alpha-mannosidase, ZINC ION | | Authors: | Santos, C.R, Cordeiro, R.L, Domingues, M.N, Borges, A.C, de Farias, M.A, Van Heel, M, Murakami, M.T, Portugal, R.V. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structure of Bl_Man38B at 3.4 A
Nat.Chem.Biol., 2022
|
|
7JPS
 
 | |
7JWV
 
 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | Descriptor: | 5-[4-(hydroxymethyl)phenyl]-1-methyl-6-{[(1R)-1-phenylethyl]sulfanyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Hurley, T.D, Buchman, C. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
7BDW
 
 | | Crystal structure of mycobacterial PptAb in complex with ACP and compound 8918 | | Descriptor: | COENZYME A, MANGANESE (II) ION, N-(2,6-diethylphenyl)-N'-(N-ethylcarbamimidoyl)urea, ... | | Authors: | Carivenc, C, Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2020-12-22 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Phosphopantetheinyl transferase binding and inhibition by amidino-urea and hydroxypyrimidinethione compounds.
Sci Rep, 11, 2021
|
|
7JY7
 
 | | Structure of a 12 base pair RecA-D loop complex | | Descriptor: | DNA (27-MER), DNA (48-MER), MAGNESIUM ION, ... | | Authors: | Pavletich, N.P. | | Deposit date: | 2020-08-29 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of strand exchange from RecA-DNA synaptic and D-loop structures.
Nature, 586, 2020
|
|
5LI6
 
 | | Crystal structure of Mycobacterium tuberculosis CYP126A1 in complex with N-isopropyl-N-((3-(4-methoxyphenyl)-1,2,4-oxadiazol-5-yl)methyl)-2-(4-nitrophenyl)acetamide | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 126, ~{N}-[[3-(4-methoxyphenyl)-1,2,4-oxadiazol-5-yl]methyl]-2-(4-nitrophenyl)-~{N}-propan-2-yl-ethanamide | | Authors: | Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2016-07-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Characterization and Ligand/Inhibitor Identification Provide Functional Insights into the Mycobacterium tuberculosis Cytochrome P450 CYP126A1.
J. Biol. Chem., 292, 2017
|
|
5LIN
 
 | |
7JSL
 
 | |
5LDW
 
 | | Structure of mammalian respiratory Complex I, class1 | | Descriptor: | Acyl carrier protein, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Vinothkumar, K.R, Zhu, J, Hirst, J. | | Deposit date: | 2016-06-28 | | Release date: | 2016-09-07 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structure of mammalian respiratory complex I.
Nature, 536, 2016
|
|
8SP9
 
 | |
8TJ2
 
 | | CryoEM structure of Myxococcus xanthus type IV pilus | | Descriptor: | Type IV major pilin protein PilA | | Authors: | Zheng, W, Egelman, E.H. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Tight-packing of large pilin subunits provides distinct structural and mechanical properties for the Myxococcus xanthus type IVa pilus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4ZVQ
 
 | |
