6ZW1
 
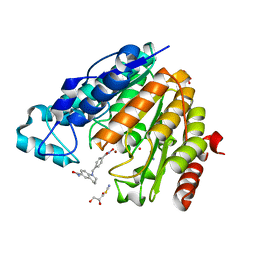 | | X-ray structure of Danio rerio histone deacetylase 6 (HDAC6) CD2 in complex with an inhibitor SW101 | | Descriptor: | 1-[[4-(oxidanylcarbamoyl)phenyl]methyl]-3,4-dihydro-2~{H}-quinoline-6-carboxamide, GLYCEROL, Histone deacetylase 6, ... | | Authors: | Barinka, C, Motlova, L, Ustinova, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Tetrahydroquinoline-Capped Histone Deacetylase 6 Inhibitor SW-101 Ameliorates Pathological Phenotypes in a Charcot-Marie-Tooth Type 2A Mouse Model.
J.Med.Chem., 64, 2021
|
|
3K8P
 
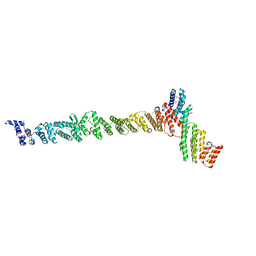 | |
4TSS
 
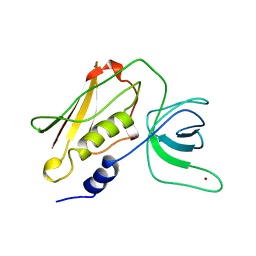 | | TOXIC SHOCK SYNDROME TOXIN-1: TETRAGONAL P4(1)2(1)2 CRYSTAL FORM | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1, ZINC ION | | Authors: | Prasad, G.S, Radhakrishnan, R, Mitchell, D.T, Earhart, C.A, Dinges, M.M, Cook, W.J, Schlivert, P.M, Ohlendorf, D.H. | | Deposit date: | 1996-12-11 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Refined structures of three crystal forms of toxic shock syndrome toxin-1 and of a tetramutant with reduced activity.
Protein Sci., 6, 1997
|
|
6J3M
 
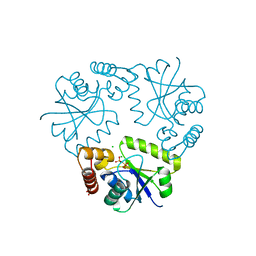 | | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Pyrophosphate at 2.30A resolution | | Descriptor: | CHLORIDE ION, PYROPHOSPHATE 2-, Phosphopantetheine adenylyltransferase, ... | | Authors: | Singh, P.K, Gupta, A, Sharma, S, Singh, T.P. | | Deposit date: | 2019-01-04 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Pyrophosphate at 2.30A resolution
To Be Published
|
|
4JNZ
 
 | |
4N00
 
 | | Discovery of 7-THP chromans: BACE1 inhibitors that reduce A-beta in the CNS | | Descriptor: | (4R,4a'S,10a'S)-2-amino-8'-(2-fluoropyridin-3-yl)-1-methyl-3',4',4a',10a'-tetrahydro-1'H-spiro[imidazole-4,10'-pyrano[4,3-b]chromen]-5(1H)-one, Beta-secretase 1, NICKEL (II) ION | | Authors: | Vigers, G.P.A, Smith, D. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of 7-tetrahydropyran-2-yl chromans: beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors that reduce amyloid beta-protein (A beta ) in the central nervous system.
J.Med.Chem., 57, 2014
|
|
4N9A
 
 | | E. coli sliding clamp in complex with (R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid | | Descriptor: | (1R)-6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-10-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach.
J.Med.Chem., 57, 2014
|
|
5X1N
 
 | | Vanillate/3-O-methylgallate O-demethylase, LigM, protocatechuate-tetrahydrofolate complex form | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Harada, A, Senda, T. | | Deposit date: | 2017-01-26 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a new O-demethylase from Sphingobium sp. strain SYK-6
FEBS J., 284, 2017
|
|
6DDI
 
 | | Crystal Structure of the human BRD2 BD1 bromodomain in complex with a Tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2S,4R)-1-acetyl-2-methyl-6-(1H-pyrazol-3-yl)-1,2,3,4-tetrahydroquinolin-4-yl]amino}benzonitrile, Bromodomain-containing protein 2, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2018-05-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bromodomain-Selective BET Inhibitors Are Potent Antitumor Agents against MYC-Driven Pediatric Cancer.
Cancer Res., 80, 2020
|
|
4ITE
 
 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with 1alpha,25-Dihydroxy-2alpha-[2-(2H-tetrazol-2-yl)ethyl]vitamin D3 | | Descriptor: | (1R,2S,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-7a-methyl-1-[(2R)-6-methyl-6-oxidanyl-heptan-2-yl]-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-2-[2-(1,2,3,4-tetrazol-2-yl)ethyl]cyclohexane-1,3-diol, Vitamin D3 receptor | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2013-01-18 | | Release date: | 2014-01-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Synthesis of 2 alpha-heteroarylalkyl active vitamin d3 with therapeutic effect on enhancing bone mineral density in vivo
ACS MED.CHEM.LETT., 4, 2013
|
|
6KB1
 
 | | X-ray structure of human PPARalpha ligand binding domain-tetradecylthioacetic acid (TTA) co-crystals obtained by soaking | | Descriptor: | 2-tetradecylsulfanylethanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
4ITL
 
 | | Crystal structure of LpxK from Aquifex aeolicus in complex with AMP-PCP at 2.1 angstrom resolution | | Descriptor: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Tetraacyldisaccharide 4'-kinase | | Authors: | Emptage, R.P, Pemble IV, C.W, York, J.D, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Mechanistic Characterization of the Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK Involved in Lipid A Biosynthesis.
Biochemistry, 52, 2013
|
|
5X1M
 
 | | Vanillate/3-O-methylgallate O-demethylase, LigM, protocatechuate-tetrahydrofolate complex form | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Harada, A, Senda, T. | | Deposit date: | 2017-01-26 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a new O-demethylase from Sphingobium sp. strain SYK-6
FEBS J., 284, 2017
|
|
6SR4
 
 | | X-ray pump X-ray probe on lysozyme.Gd nanocrystals: 112 fs time delay | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Kloos, M, Gorel, A, Nass, K. | | Deposit date: | 2019-09-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural dynamics in proteins induced by and probed with X-ray free-electron laser pulses.
Nat Commun, 11, 2020
|
|
6SR3
 
 | | X-ray pump X-ray probe on lysozyme.Gd nanocrystals: 62 fs time delay | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Kloos, M, Gorel, A, Nass, K. | | Deposit date: | 2019-09-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural dynamics in proteins induced by and probed with X-ray free-electron laser pulses.
Nat Commun, 11, 2020
|
|
6SR0
 
 | | X-ray pump X-ray probe on lysozyme.Gd nanocrystals: single colour reference data | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Kloos, M, Gorel, A, Nass, K. | | Deposit date: | 2019-09-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural dynamics in proteins induced by and probed with X-ray free-electron laser pulses.
Nat Commun, 11, 2020
|
|
5X4A
 
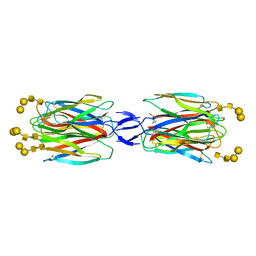 | | SLL-2-Forssman antigen tetrasaccharides complex | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Galactose-binding lectin, ... | | Authors: | Kita, A, Miki, K. | | Deposit date: | 2017-02-11 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of octocoral lectin SLL-2 complexed with Forssman antigen tetrasaccharide.
Glycobiology, 2017
|
|
2OXX
 
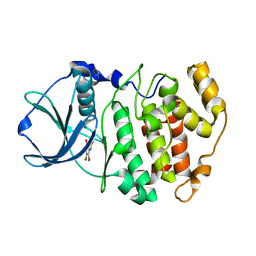 | | Protein kinase CK2 in complex with tetrabromobenzoimidazole derivatives K17, K22 and K32 | | Descriptor: | 4,5,6,7-TETRABROMO-1H,3H-BENZIMIDAZOL-2-THIONE, Casein kinase II subunit alpha | | Authors: | Battistutta, R, Zanotti, G, Cendron, L. | | Deposit date: | 2007-02-21 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The ATP-Binding Site of Protein Kinase CK2 Holds a Positive Electrostatic Area and Conserved Water Molecules.
Chembiochem, 8, 2007
|
|
2OXY
 
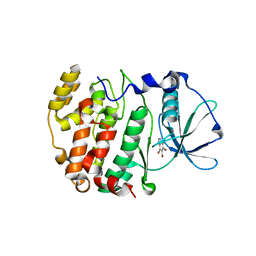 | | Protein kinase CK2 in complex with tetrabromobenzoimidazole derivatives K17, K22 and K32 | | Descriptor: | 4,5,6,7-TETRABROMO-BENZIMIDAZOLE, Casein kinase II subunit alpha | | Authors: | Battistutta, R, Zanotti, G, Cendron, L. | | Deposit date: | 2007-02-21 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.812 Å) | | Cite: | The ATP-Binding Site of Protein Kinase CK2 Holds a Positive Electrostatic Area and Conserved Water Molecules.
Chembiochem, 8, 2007
|
|
5GY0
 
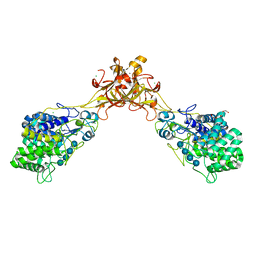 | | Crystal structure of endoglucanase CelQ from Clostridium thermocellum complexed with cellotetraose | | Descriptor: | BROMIDE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2016-09-21 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structures of the C-Terminally Truncated Endoglucanase Cel9Q from Clostridium thermocellum Complexed with Cellodextrins and Tris.
Chembiochem, 20, 2019
|
|
3ALB
 
 | | Cyclic Lys48-linked tetraubiquitin | | Descriptor: | SULFATE ION, ubiquitin | | Authors: | Satoh, T, Sakata, E, Yamamoto, S, Yamaguchi, Y, Sumiyoshi, A, Wakatsuki, S, Kato, K. | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of cyclic Lys48-linked tetraubiquitin
Biochem.Biophys.Res.Commun., 2010
|
|
3N3I
 
 | | Crystal Structure of G48V/C95F tethered HIV-1 Protease/Saquinavir complex | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Protease | | Authors: | Prashar, V, Bihani, S.C, Das, A, Rao, D.R, Hosur, M.V. | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Insights into the mechanism of drug resistance: X-ray structure analysis of G48V/C95F tethered HIV-1 protease dimer/saquinavir complex
Biochem.Biophys.Res.Commun., 396, 2010
|
|
4PNU
 
 | | E. coli sliding clamp in complex with (R)-6-bromo-9-(2-((R)-1-carboxy-2-phenylethylamino)-2-oxoethyl)-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid | | Descriptor: | (2R)-6-bromo-9-(2-{[(1R)-1-carboxy-2-phenylethyl]amino}-2-oxoethyl)-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacterial Sliding Clamp Inhibitors that Mimic the Sequential Binding Mechanism of Endogenous Linear Motifs.
J.Med.Chem., 58, 2015
|
|
4ISH
 
 | | Structure of FACTOR VIIA in complex with the inhibitor BMS-593214 also known as 2'-[(6R,6AR,11BR)-2-CARBAMIMIDOYL-6,6A,7,11B-TETRAHYDRO-5H-INDENO[2,1-C]QUINOLIN-6-YL]-5'-HYDROXY-4'-METHOXYBIPHENYL-4-CARBOXYLIC ACID | | Descriptor: | 2'-[(6R,6aR,11bR)-2-carbamimidoyl-6,6a,7,11b-tetrahydro-5H-indeno[2,1-c]quinolin-6-yl]-5'-hydroxy-4'-methoxybiphenyl-4-carboxylic acid, CALCIUM ION, Factor VII heavy chain, ... | | Authors: | Wei, A. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery and gram-scale synthesis of BMS-593214, a potent, selective FVIIa inhibitor.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7KXJ
 
 | | SARS-CoV-2 spike protein in complex with Fab 15033-7, 3-"up", asymmetric | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J. | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-03 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
