9B8Z
 
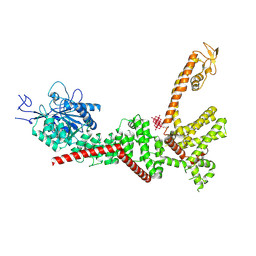 | |
8XUV
 
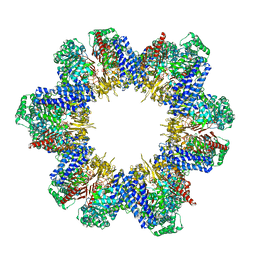 | | Cryo-EM structure of tomato NRC2 filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, NRC2 | | Authors: | Sun, Y, Ma, S.C, Chai, J.J. | | Deposit date: | 2024-01-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Oligomerization-mediated autoinhibition and cofactor binding of a plant NLR.
Nature, 2024
|
|
9ASB
 
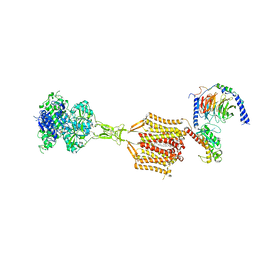 | | Structure of human calcium-sensing receptor in complex with chimeric Gq (miniGisq) protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-02-24 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
8URV
 
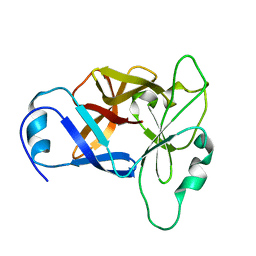 | |
9B94
 
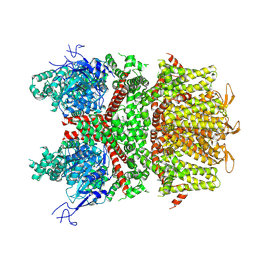 | |
8USX
 
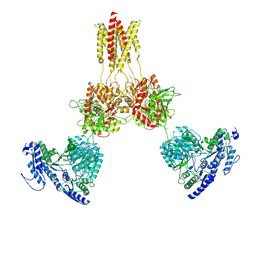 | | Glycine-bound GluN1a-3A NMDA receptor | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 3A | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure and function of GluN1-3A NMDA receptor excitatory glycine receptor channel.
Sci Adv, 10, 2024
|
|
9AVL
 
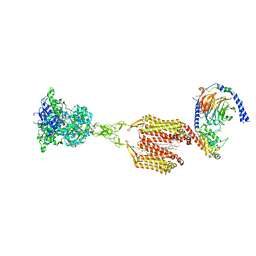 | | Structure of human calcium-sensing receptor in complex with Gi3 protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-04 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
8Y90
 
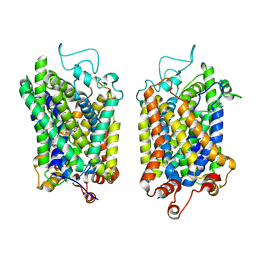 | | Structure of NET-Nefopam in outward-open state | | Descriptor: | (1S)-5-methyl-1-phenyl-1,3,4,6-tetrahydro-2,5-benzoxazocine, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
9B90
 
 | |
9B93
 
 | |
8WGK
 
 | |
8WPF
 
 | | Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with exogenous DNA bearing one abasic site | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, A22R DNA polymerase processivity factor, DNA polymerase, ... | | Authors: | Wang, X, Li, N, Gao, N. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the assembly and mechanism of mpox virus DNA polymerase complex F8-A22-E4-H5.
Mol.Cell, 83, 2023
|
|
8Y92
 
 | | structure of NET-Atomoxetine in outward-open state | | Descriptor: | (3R)-N-methyl-3-(2-methylphenoxy)-3-phenyl-propan-1-amine, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
9EM1
 
 | | Human pyridoxal phosphatase in complex with 7,8-dihydroxyflavone and phosphate | | Descriptor: | 7,8-bis(oxidanyl)-2-phenyl-chromen-4-one, Chronophin, GLYCEROL, ... | | Authors: | Brenner, M, Gohla, A, Schindelin, H. | | Deposit date: | 2024-03-07 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 7,8-Dihydroxyflavone is a direct inhibitor of human and murine pyridoxal phosphatase.
Elife, 13, 2024
|
|
8Y93
 
 | | Structure of NET-Amitriptyline in outward-open state | | Descriptor: | Amitriptyline, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8WPP
 
 | | Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with endogenous DNA | | Descriptor: | A22R DNA polymerase processivity factor, DNA polymerase, E4R Uracil-DNA glycosylase, ... | | Authors: | Wang, X, Li, N, Gao, N. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the assembly and mechanism of mpox virus DNA polymerase complex F8-A22-E4-H5.
Mol.Cell, 83, 2023
|
|
8WMF
 
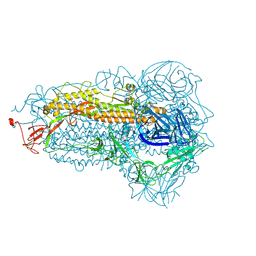 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein (closed-1 state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | Deposit date: | 2023-10-03 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 2024
|
|
8VJJ
 
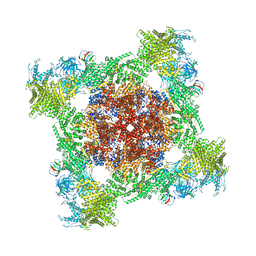 | | Structure of mouse RyR1 (EGTA-only dataset) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ... | | Authors: | Weninger, G, Marks, A.R. | | Deposit date: | 2024-01-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural insights into the regulation of RyR1 by S100A1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8WPK
 
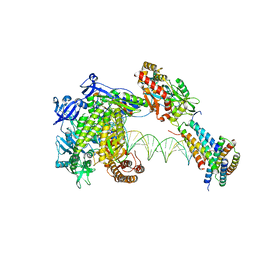 | | Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with exgenous DNA | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA polymerase, DNA polymerase processivity factor, ... | | Authors: | Wang, X, Li, N, Gao, N. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the assembly and mechanism of mpox virus DNA polymerase complex F8-A22-E4-H5.
Mol.Cell, 83, 2023
|
|
8TN1
 
 | |
8YRC
 
 | | Chlorinated YabJ from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, OXYGEN ATOM, SODIUM ION, ... | | Authors: | Jeong, C, Kim, H.J. | | Deposit date: | 2024-03-21 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | YabJ from Staphylococcus aureus entraps chlorides within its pocket.
Biochem.Biophys.Res.Commun., 710, 2024
|
|
8VQ4
 
 | | CDK2-CyclinE1 in complex with allosteric inhibitor I-125A. | | Descriptor: | (8R)-6-(1-benzyl-1H-pyrazole-4-carbonyl)-N-[(2S,3R)-3-(2-cyclohexylethoxy)-1-(methylamino)-1-oxobutan-2-yl]-2-[(1S)-2,2-dimethylcyclopropane-1-carbonyl]-2,6-diazaspiro[3.4]octane-8-carboxamide, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Hirschi, M, Johnson, E, Zhang, Y, Liu, Z, Brodsky, O, Won, S.J, Nagata, A, Petroski, M.D, Majmudar, J.D, Niessen, S, VanArsdale, T, Gilbert, A.M, Hayward, M.M, Stewart, A.E, Nager, A.R, Melillo, B, Cravatt, B. | | Deposit date: | 2024-01-17 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expanding the ligandable proteome by paralog hopping with covalent probes.
Biorxiv, 2024
|
|
8W4V
 
 | | Crystal structure of human HSP90 in complex with compound 4 | | Descriptor: | 4-[2-[(dimethylamino)methyl]phenyl]sulfanylbenzene-1,3-diol, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Xu, C, Zhang, X.L, Bai, F. | | Deposit date: | 2023-08-25 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Accurate Characterization of Binding Kinetics and Allosteric Mechanisms for the HSP90 Chaperone Inhibitors Using AI-Augmented Integrative Biophysical Studies.
Jacs Au, 4, 2024
|
|
8WZI
 
 | | One RBD up state of Spike glycoprotein, SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8VK4
 
 | |
