5DKQ
 
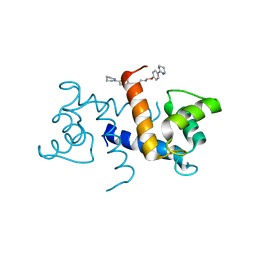 | | Crystal Structure of Calcium-loaded S100B bound to SBi4214 | | Descriptor: | 2,2'-[pentane-1,5-diylbis(oxybenzene-4,1-diyl)]di-1,4,5,6-tetrahydropyrimidine, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
5U0T
 
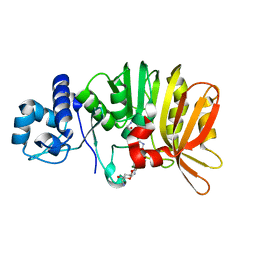 | | Crystal structure of a methyltransferase involved in the biosynthesis of gentamicin in complex with (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-3-(methylamino)-alpha-D-glucopyranosyl]oxy}-2-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-3-(methylamino)-alpha-D-glucopyranosyl]oxy}-2-hydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, MAGNESIUM ION, Putative gentamicin methyltransferase, ... | | Authors: | Bury, P, Huang, F, Leadlay, P, Dias, M.V.B. | | Deposit date: | 2016-11-27 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Structural Basis of the Selectivity of GenN, an Aminoglycoside N-Methyltransferase Involved in Gentamicin Biosynthesis.
ACS Chem. Biol., 12, 2017
|
|
6G3Y
 
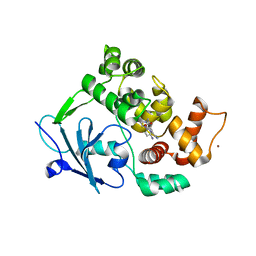 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH5675 | | Descriptor: | 4-(4-azanyl-2-oxidanylidene-3~{H}-benzimidazol-1-yl)-~{N}-(4-iodophenyl)piperidine-1-carboxamide, ACETATE ION, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Helleday, T, Stenmark, P. | | Deposit date: | 2018-03-26 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation.
Science, 362, 2018
|
|
5KYE
 
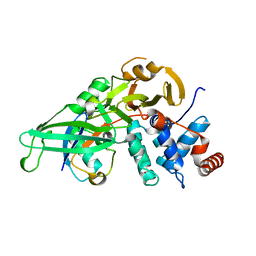 | |
6PR9
 
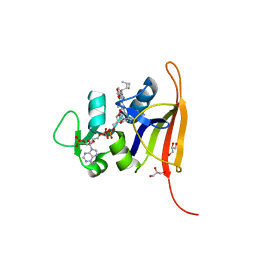 | | S. aureus dihydrofolate reductase co-crystallized with 1-ethylpropyl-dihydropthalazine inhibitor and NADP(H) | | Descriptor: | (2E)-3-{5-[(2,4-diaminopyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1S)-1-(pentan-3-yl)-3,4-dihydrophthalazin-2(1H)-yl]prop-2-en-1-one, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Bourne, C.R, Thomas, L.M. | | Deposit date: | 2019-07-10 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Inhibitor design to target a unique feature in the folate pocket of Staphylococcus aureus dihydrofolate reductase.
Eur.J.Med.Chem., 200, 2020
|
|
6FNR
 
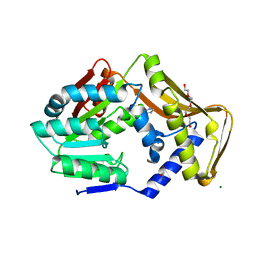 | | Ergothioneine-biosynthetic methyltransferase EgtD in complex with chlorohistidine | | Descriptor: | (2~{S})-2-chloranyl-3-(1~{H}-imidazol-5-yl)propanoic acid, GLYCEROL, Histidine N-alpha-methyltransferase, ... | | Authors: | Vit, A, Blankenfeldt, W, Seebeck, F.P. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Inhibition and Regulation of the Ergothioneine Biosynthetic Methyltransferase EgtD.
ACS Chem. Biol., 13, 2018
|
|
6YP8
 
 | |
6MH9
 
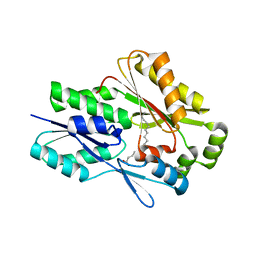 | | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein A121I mutant to 2.02 Angstrom resolution | | Descriptor: | Fatty Acid Kinase (Fak) B1 protein, PALMITIC ACID | | Authors: | Cuypers, M.G, Ericson, M, Subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2018-09-17 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification of structural transitions in bacterial fatty acid binding proteins that permit ligand entry and exit at membranes.
J.Biol.Chem., 298, 2022
|
|
6YPD
 
 | | Crystal structure of AmpC from E. coli with Cyclic Boronate 3 (CB3 / APC308) | | Descriptor: | (3~{S})-2,2-bis(oxidanyl)-3-(phenylmethylsulfanyl)-3,4-dihydro-1,2-benzoxaborinin-2-ium-8-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Lang, P.A, Brem, J, Schofield, C.J. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bicyclic Boronates as Potent Inhibitors of AmpC, the Class C beta-Lactamase from Escherichia coli .
Biomolecules, 10, 2020
|
|
5U0Z
 
 | |
6YQ3
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | (3~{R})-8-methoxy-3-methyl-3,6-bis(oxidanyl)-2,4-dihydrobenzo[a]anthracene-1,7,12-trione, 1,2-ETHANEDIOL, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
5U17
 
 | |
5L1O
 
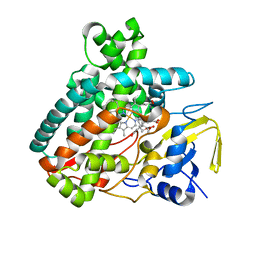 | | X-ray Structure of Cytochrome P450 PntM with Pentalenolactone F | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase, pentalenolactone F | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
6G4Q
 
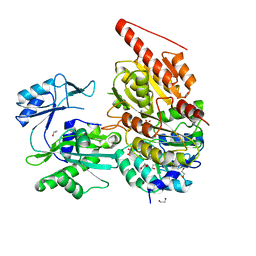 | | Structure of human ADP-forming succinyl-CoA ligase complex SUCLG1-SUCLA2 | | Descriptor: | 1,2-ETHANEDIOL, Succinate--CoA ligase [ADP-forming] subunit beta, mitochondrial, ... | | Authors: | Bailey, H.J, Shrestha, L, Rembeza, E, Sorrell, F.J, Newman, J, Strain-Damerell, C, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2018-03-28 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of human ADP-forming succinyl-CoA ligase complex SUCLG1-SUCLA2
To Be Published
|
|
8OT8
 
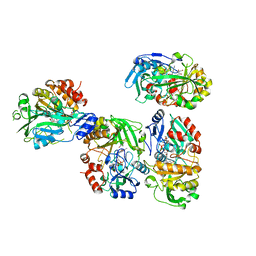 | |
7DJQ
 
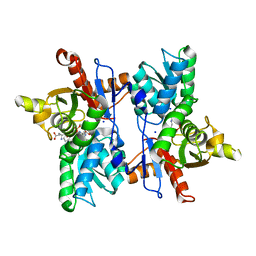 | |
5FHH
 
 | | Structure of human Pif1 helicase domain residues 200-641 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase PIF1, TETRAFLUOROALUMINATE ION | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
5L7G
 
 | | MCR IN COMPLEX WITH ligand | | Descriptor: | 1,2-ETHANEDIOL, Mineralocorticoid receptor, NCOA1 peptide, ... | | Authors: | Edman, K, Aagaard, A, Backstrom, S, Xue, Y. | | Deposit date: | 2016-06-03 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Drug Design of Mineralocorticoid Receptor Antagonists to Explore Oxosteroid Receptor Selectivity.
ChemMedChem, 12, 2017
|
|
6GAM
 
 | | Structure of E14Q variant of E. coli hydrogenase-2 (as-isolated enzyme) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-04-11 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
5L7N
 
 | | Plexin A1 extracellular fragment, domains 7-10 (IPT3-IPT6) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kong, Y, Janssen, B.J.C, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5U2V
 
 | |
6YQN
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual inhibitor TW9 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-4-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]t rideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]benzamide | | Authors: | Joerger, A.C, Balourdas, D.I, Weiser, T, Chatterjee, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Characterization of a dual BET/HDAC inhibitor for treatment of pancreatic ductal adenocarcinoma.
Int.J.Cancer, 147, 2020
|
|
6G54
 
 | | Crystal structure of ERK2 covalently bound to SM1-71 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Mitogen-activated protein kinase 1, ... | | Authors: | Chaikuad, A, Suman, R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-29 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Leveraging Compound Promiscuity to Identify Targetable Cysteines within the Kinome.
Cell Chem Biol, 26, 2019
|
|
6V6Q
 
 | | Crystal Structure of Monophosphorylated FGF Receptor 2 isoform IIIb with PTR657 | | Descriptor: | Fibroblast growth factor receptor 2, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Lin, C.-C, Wieteska, L, Poncet-Montange, G, Suen, K.M, Arold, S.T, Ahmed, Z, Ladbury, J.E. | | Deposit date: | 2019-12-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The combined action of the intracellular regions regulates FGFR2 kinase activity
Commun Biol, 6, 2023
|
|
7DM2
 
 | | crystal structure of the M. tuberculosis phosphate ABC transport receptor PstS-1 in complex with Fab p4-170 | | Descriptor: | PHOSPHATE ION, Phosphate-binding protein PstS 1, heavy chain, ... | | Authors: | Ma, B, Freund, N, Xiang, Y. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human antibodies targeting a Mycobacterium transporter protein mediate protection against tuberculosis.
Nat Commun, 12, 2021
|
|
