3I4B
 
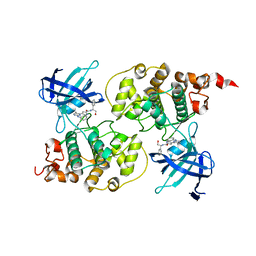 | | Crystal structure of GSK3b in complex with a pyrimidylpyrrole inhibitor | | Descriptor: | Glycogen synthase kinase-3 beta, N-[(1S)-2-hydroxy-1-phenylethyl]-4-[5-methyl-2-(phenylamino)pyrimidin-4-yl]-1H-pyrrole-2-carboxamide | | Authors: | Ter Haar, E. | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of potent and selective pyrimidylpyrrole inhibitors of extracellular signal-regulated kinase (ERK) using conformational control.
J.Med.Chem., 52, 2009
|
|
3HV0
 
 | |
3I4Z
 
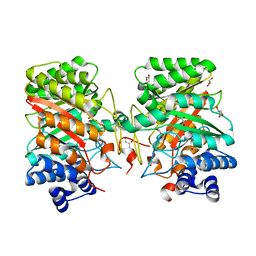 | | Crystal structure of the dimethylallyl tryptophan synthase FgaPT2 from Aspergillus fumigatus | | Descriptor: | 1,3-BUTANEDIOL, GLYCEROL, Tryptophan dimethylallyltransferase | | Authors: | Schall, C, Zocher, G, Stehle, T. | | Deposit date: | 2009-07-03 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3PUI
 
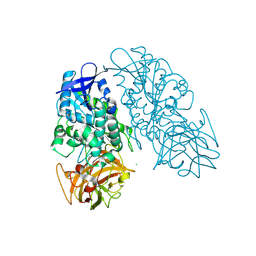 | | Cocaine Esterase with mutations G4C, S10C | | Descriptor: | CHLORIDE ION, Cocaine esterase, SODIUM ION | | Authors: | Nance, M.R, Tesmer, J.J.G. | | Deposit date: | 2010-12-04 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Subunit stabilization and polyethylene glycolation of cocaine esterase improves in vivo residence time.
Mol.Pharmacol., 80, 2011
|
|
3I57
 
 | | Type 2 repeat of the mucus binding protein MUB from Lactobacillus reuteri | | Descriptor: | CALCIUM ION, Mucus binding protein | | Authors: | Hemmings, A.M, MacKenzie, D.A, Tailford, L.E, Juge, N. | | Deposit date: | 2009-07-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a mucus binding protein repeat reveals an unexpected functional immunoglobulin binding activity.
J.Biol.Chem., 284, 2009
|
|
3PVU
 
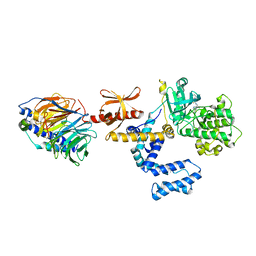 | | Bovine GRK2 in complex with Gbetagamma subunits and a selective kinase inhibitor (CMPD101) | | Descriptor: | 3-({[4-methyl-5-(pyridin-4-yl)-4H-1,2,4-triazol-3-yl]methyl}amino)-N-[2-(trifluoromethyl)benzyl]benzamide, Beta-adrenergic receptor kinase 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Thal, D.M, Tesmer, J.J. | | Deposit date: | 2010-12-07 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Molecular Mechanism of Selectivity among G Protein-Coupled Receptor Kinase 2 Inhibitors.
Mol.Pharmacol., 80, 2011
|
|
3HWI
 
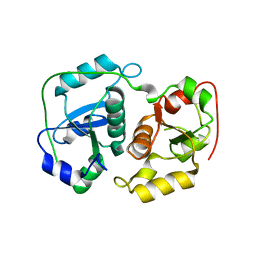 | |
3PWD
 
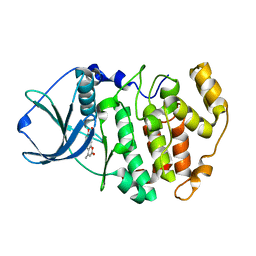 | | Crystal structure of maize CK2 in complex with NBC (Z1) | | Descriptor: | 8-hydroxy-4-methyl-9-nitro-2H-benzo[g]chromen-2-one, Casein kinase II subunit alpha | | Authors: | Battistutta, R, Mazzorana, M. | | Deposit date: | 2010-12-08 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural features underlying the selectivity of the kinase inhibitors NBC and dNBC: role of a nitro group that discriminates between CK2 and DYRK1A
Cell.Mol.Life Sci., 69, 2012
|
|
3PR6
 
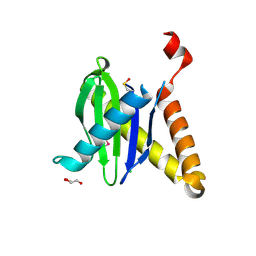 | |
3HML
 
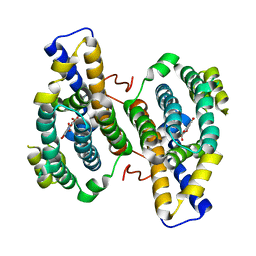 | |
3PRD
 
 | |
3HX3
 
 | | Crystal structure of CRALBP mutant R234W | | Descriptor: | RETINAL, Retinaldehyde-binding protein 1 | | Authors: | Stocker, A, He, X, Lobsiger, J. | | Deposit date: | 2009-06-19 | | Release date: | 2009-10-20 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Bothnia dystrophy is caused by domino-like rearrangements in cellular retinaldehyde-binding protein mutant R234W.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3PWL
 
 | | Human Class I MHC HLA-A2 in complex with the HuD peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Borbulevych, O.Y, Baker, B.M. | | Deposit date: | 2010-12-08 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Conformational Melding Permits a Conserved Binding Geometry in TCR Recognition of Foreign and Self Molecular Mimics.
J.Immunol., 186, 2011
|
|
3PX6
 
 | |
3GLP
 
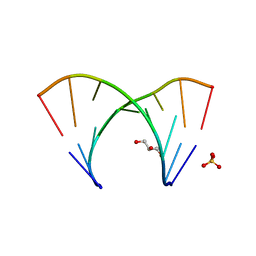 | | 1.23 A resolution X-ray structure of (GCUGCUGC)2 | | Descriptor: | 5'-R(*GP*CP*UP*GP*CP*UP*GP*C)-3', GLYCEROL, SULFATE ION | | Authors: | Kiliszek, A, Kierzek, R, Krzyzosiak, W.J, Rypniewski, W. | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structural insights into CUG repeats containing the 'stretched U-U wobble': implications for myotonic dystrophy.
Nucleic Acids Res., 37, 2009
|
|
3PZZ
 
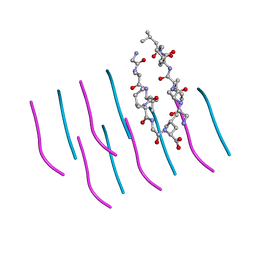 | |
3H85
 
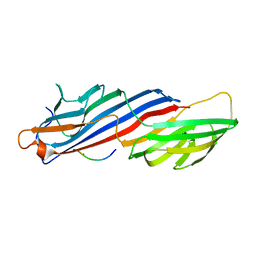 | | Molecular basis for the association of PIPKI gamma-p90 with the clathrin adaptor AP-2 | | Descriptor: | AP-2 complex subunit mu-1, NICKEL (II) ION, Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma | | Authors: | Vahedi-Faridi, A, Kahlfeldt, N, Schaefer, J.G, Haucke, V. | | Deposit date: | 2009-04-28 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for association of PIPKI gamma-p90 with clathrin adaptor AP-2.
J.Biol.Chem., 285, 2010
|
|
3Q0X
 
 | | N-terminal coiled-coil dimer domain of C. reinhardtii SAS-6 homolog Bld12p | | Descriptor: | Centriole protein | | Authors: | Kitagawa, D, Vakonakis, I, Olieric, N, Hilbert, M, Keller, D, Olieric, V, Bortfeld, M, Erat, M.C, Flueckiger, I, Goenczy, P, Steinmetz, M.O. | | Deposit date: | 2010-12-16 | | Release date: | 2011-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis of the 9-fold symmetry of centrioles.
Cell(Cambridge,Mass.), 144, 2011
|
|
3H8G
 
 | | Bestatin complex structure of leucine aminopeptidase from Pseudomonas putida | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, BICARBONATE ION, Cytosol aminopeptidase, ... | | Authors: | Kale, A, Dijkstra, B.W, Sonke, T, Thunnissen, A.M.W.H. | | Deposit date: | 2009-04-29 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the leucine aminopeptidase from Pseudomonas putida reveals the molecular basis for its enantioselectivity and broad substrate specificity.
J.Mol.Biol., 398, 2010
|
|
3PU4
 
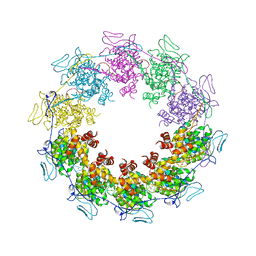 | |
3PV8
 
 | |
3GMV
 
 | | Crystal Structure of Beta-Lactamse Inhibitory Protein-I (BLIP-I) in Apo Form | | Descriptor: | Beta-lactamase inhibitory protein BLIP-I, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Lim, D.C, Gretes, M, Strynadka, N.C.J. | | Deposit date: | 2009-03-15 | | Release date: | 2009-03-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
3HAF
 
 | | Human prion protein variant V129 domain swapped dimer | | Descriptor: | CADMIUM ION, CHLORIDE ION, Major prion protein | | Authors: | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | Deposit date: | 2009-05-01 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
3Q2S
 
 | | Crystal Structure of CFIm68 RRM/CFIm25 complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 5, Cleavage and polyadenylation specificity factor subunit 6 | | Authors: | Yang, Q, Coseno, M, Gilmartin, G.M, Doublie, S. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Human Cleavage Factor CFI(m)25/CFI(m)68/RNA Complex Provides an Insight into Poly(A) Site Recognition and RNA Looping.
Structure, 19, 2011
|
|
3Q4J
 
 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
