3OF8
 
 | |
3OFE
 
 | |
3OEO
 
 | | The crystal structure E. coli Spy | | Descriptor: | CADMIUM ION, Spheroplast protein Y | | Authors: | Kwon, E, Kim, D.Y, Gross, C.A, Gross, J.D, Kim, K.K. | | Deposit date: | 2010-08-13 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure Escherichia coli Spy.
Protein Sci., 19, 2010
|
|
3OG6
 
 | | The crystal structure of human interferon lambda 1 complexed with its high affinity receptor in space group P212121 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Interleukin 28 receptor, ... | | Authors: | Miknis, Z.J, Magracheva, E, Lei, W, Zdanov, A, Kotenko, S.V, Wlodawer, A. | | Deposit date: | 2010-08-16 | | Release date: | 2010-10-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of the complex of human interferon-lambda1 with its high affinity receptor interferon-lambdaR1.
J.Mol.Biol., 404, 2010
|
|
3OGC
 
 | | KcsA E71A variant in presence of Na+ | | Descriptor: | SODIUM ION, Voltage-gated potassium channel, antibody Fab fragment heavy chain, ... | | Authors: | McCoy, J.G, Nimigean, C.M. | | Deposit date: | 2010-08-16 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Mechanism for selectivity-inactivation coupling in KcsA potassium channels.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OFG
 
 | | Structured Domain of Caenorhabditis elegans BMY-1 | | Descriptor: | Boca/mesd chaperone for ywtd beta-propeller-egf protein 1, CHLORIDE ION | | Authors: | Collins, M.N, Hendrickson, W.A. | | Deposit date: | 2010-08-14 | | Release date: | 2011-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.367 Å) | | Cite: | Structural Characterization of the Boca/Mesd Maturation Factors for LDL-Receptor-Type beta-Propeller Domains
Structure, 2011
|
|
3OAU
 
 | | Antibody 2G12 Recognizes Di-Mannose Equivalently in Domain- and Non-Domain-Exchanged Forms, but only binds the HIV-1 Glycan Shield if Domain-Exchanged | | Descriptor: | Fab 2G12, heavy chain, light chain, ... | | Authors: | Doores, K.J, Fulton, Z, Huber, M, Wilson, I.A, Burton, D.R. | | Deposit date: | 2010-08-05 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody 2G12 recognizes di-mannose equivalently in domain- and nondomain-exchanged forms but only binds the HIV-1 glycan shield if domain exchanged.
J.Virol., 84, 2010
|
|
3OGD
 
 | | AlkA Undamaged DNA Complex: Interrogation of a G*:C base pair | | Descriptor: | 5'-D(*CP*AP*(BRU)P*GP*AP*CP*(BRU)P*GP*C)-3', 5'-D(*GP*CP*AP*GP*TP*CP*AP*TP*G)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2010-08-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Escherichia coli AlkA in Complex with Undamaged DNA.
J.Biol.Chem., 285, 2010
|
|
3OBZ
 
 | |
3OCU
 
 | | Structure of Recombinant Haemophilus Influenzae e(P4) Acid Phosphatase mutant D66N complexed with NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Lipoprotein E, MAGNESIUM ION | | Authors: | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | Deposit date: | 2010-08-10 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Recognition of nucleoside monophosphate substrates by Haemophilus influenzae class C acid phosphatase.
J.Mol.Biol., 404, 2010
|
|
3OD4
 
 | | Crystal Structure of Factor Inhibiting HIF-1 Alpha Complexed with Inhibitor | | Descriptor: | 8-hydroxyquinoline-5-carboxylic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | King, O.N.F, Bashford-Rogers, R, Maloney, D.J, Jadhav, A, Heightman, T.D, Simeonov, A, Clifton, I.J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2010-08-10 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Factor Inhibiting HIF-1 Alpha Complexed with Inhibitor
To be Published
|
|
3ODE
 
 | | Human PARP-1 zinc finger 2 (Zn2) bound to DNA | | Descriptor: | 5'-D(*CP*CP*CP*AP*AP*GP*CP*G)-3', 5'-D(*CP*GP*CP*TP*TP*GP*GP*G)-3', Poly [ADP-ribose] polymerase 1, ... | | Authors: | Pascal, J.M, Langelier, M.-F. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structures of Poly(ADP-ribose) Polymerase-1 (PARP-1) Zinc Fingers Bound to DNA: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO DNA-DEPENDENT PARP-1 ACTIVITY.
J.Biol.Chem., 286, 2011
|
|
3OFH
 
 | | Structured Domain of Mus musculus Mesd | | Descriptor: | LDLR chaperone MESD | | Authors: | Collins, M.N, Hendrickson, W.A. | | Deposit date: | 2010-08-14 | | Release date: | 2011-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | Structural Characterization of the Boca/Mesd Maturation Factors for LDL-Receptor-Type beta-Propeller Domains
Structure, 2011
|
|
3OFS
 
 | | Dynamic conformations of the CD38-mediated NAD cyclization captured using multi-copy crystallography | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Zhang, H, Lee, H.C, Hao, Q. | | Deposit date: | 2010-08-16 | | Release date: | 2010-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamic Conformations of the CD38-Mediated NAD Cyclization Captured in a Single Crystal
J.Mol.Biol., 405, 2011
|
|
3OEU
 
 | | Structure of yeast 20S open-gate proteasome with Compound 24 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-{(2S)-1-[(2-chlorobenzyl)amino]-1-oxo-4-phenylbutan-2-yl}-N~2~-[3-(2-methylphenyl)propanoyl]-L-threoninamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-08-13 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Optimization of a series of dipeptides with a P3 threonine residue as non-covalent inhibitors of the chymotrypsin-like activity of the human 20S proteasome.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OGQ
 
 | | Crystal Structure of 6s-98S FIV Protease with Lopinavir bound | | Descriptor: | DIMETHYL SULFOXIDE, FIV protease, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE | | Authors: | Lin, Y.-C, Perryman, A.L, Elder, J.H, Stout, C.D. | | Deposit date: | 2010-08-17 | | Release date: | 2011-06-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for drug and substrate specificity exhibited by FIV encoding a chimeric FIV/HIV protease.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3OH3
 
 | | Protein structure of USP from L. major bound to URIDINE-5'-DIPHOSPHATE -Arabinose | | Descriptor: | GLYCEROL, UDP-sugar pyrophosphorylase, [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2S,3R,4S,5S)-3,4,5-trihydroxytetrahydro-2H-pyran-2-yl dihydrogen diphosphate | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
3OCR
 
 | | Crystal structure of aldolase II superfamily protein from Pseudomonas syringae | | Descriptor: | Class II aldolase/adducin domain protein, SULFATE ION | | Authors: | Chang, C, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-10 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of aldolase II superfamily protein from Pseudomonas syringae
To be Published
|
|
3OH4
 
 | | Protein structure of USP from L. major bound to URIDINE-5'-DIPHOSPHATE Glucose | | Descriptor: | GLYCEROL, UDP-sugar pyrophosphorylase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F.H, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
3ODC
 
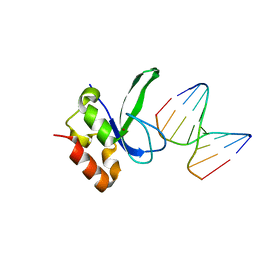 | | Human PARP-1 zinc finger 2 (Zn2) bound to DNA | | Descriptor: | 5'-D(*CP*CP*CP*AP*GP*AP*CP*G)-3', 5'-D(*CP*GP*TP*CP*TP*GP*GP*G)-3', Poly [ADP-ribose] polymerase 1, ... | | Authors: | Pascal, J.M, Langelier, M.-F. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Poly(ADP-ribose) Polymerase-1 (PARP-1) Zinc Fingers Bound to DNA: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO DNA-DEPENDENT PARP-1 ACTIVITY.
J.Biol.Chem., 286, 2011
|
|
3ODJ
 
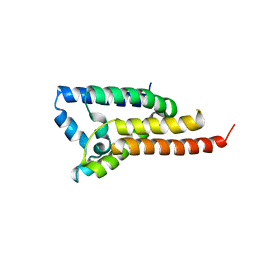 | | Crystal structure of H. influenzae rhomboid GlpG with disordered loop 4, helix 5 and loop 5 | | Descriptor: | Rhomboid protease glpG | | Authors: | Brooks, C.L, Lazareno-Saez, C, Lamoureux, J.S, Mak, M.W, Lemieux, M.J. | | Deposit date: | 2010-08-11 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Insights into Substrate Gating in H. influenzae Rhomboid.
J.Mol.Biol., 407, 2011
|
|
3OH2
 
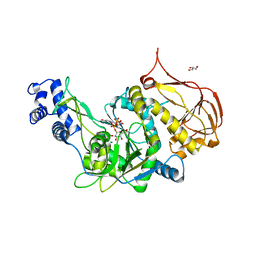 | | Protein structure of USP from L. major bound to URIDINE-5'-DIPHOSPHATE-GALACTOSE | | Descriptor: | GALACTOSE-URIDINE-5'-DIPHOSPHATE, GLYCEROL, UDP-sugar pyrophosphorylase | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
3OHH
 
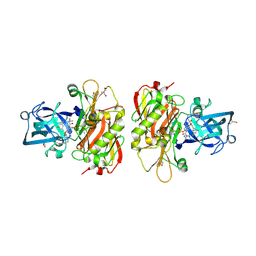 | | Crystal structure of beta-site app-cleaving enzyme 1 (bace-wt) complex with bms-681889 aka n~1~-butyl-5-cyano- n~3~-((1s,2r)-1-(3,5-difluorobenzyl)-2-hydroxy-3-((3- methoxybenzyl)amino)propyl)-n~1~-methyl-1h-indole-1,3- dicarboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, N~1~-butyl-5-cyano-N~3~-{(1S,2R)-1-(3,5-difluorobenzyl)-2-hydroxy-3-[(3-methoxybenzyl)amino]propyl}-N~1~-methyl-1H-indole-1,3-dicarboxamide, ... | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2010-08-17 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Synthesis and SAR of indole-and 7-azaindole-1,3-dicarboxamide hydroxyethylamine inhibitors of BACE-1.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3OF9
 
 | |
3OFI
 
 | | Crystal structure of human insulin-degrading enzyme in complex with ubiquitin | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Insulin-degrading enzyme, Ubiquitin, ... | | Authors: | Kalas, V, Ralat, L.A, Tang, W.-J. | | Deposit date: | 2010-08-15 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Ubiquitin is a novel substrate for human insulin-degrading enzyme.
J.Mol.Biol., 406, 2011
|
|
