7OCJ
 
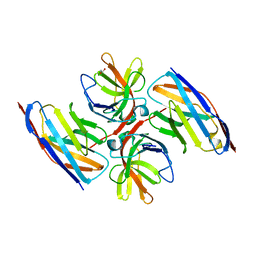 | | Crystal structure of E.coli LexA in complex with nanobody NbSOS2(Nb14509) | | Descriptor: | 1,2-ETHANEDIOL, LexA repressor, NbSOS2 (14509) | | Authors: | Maso, L, Vascon, F, Chinellato, M, Pardon, E, Steyaert, J, Angelini, A, Tondi, D, Cendron, L. | | Deposit date: | 2021-04-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies targeting LexA autocleavage disclose a novel suppression strategy of SOS-response pathway.
Structure, 30, 2022
|
|
7AAB
 
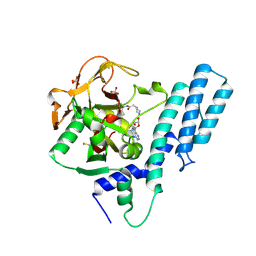 | | Crystal structure of the catalytic domain of human PARP1 in complex with inhibitor EB-47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
7AAA
 
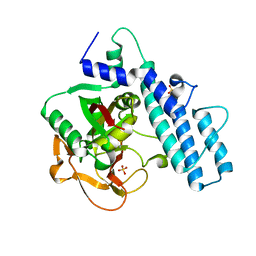 | | Crystal structure of the catalytic domain of human PARP1 (apo) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Underwood, E, Rawlins, P.B, Johannes, J.W, Easton, L.E, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
3BPV
 
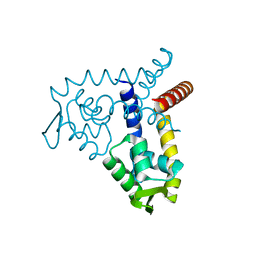 | | Crystal Structure of MarR | | Descriptor: | Transcriptional regulator | | Authors: | Saridakis, V, Shahinas, D, Xu, X, Christendat, D. | | Deposit date: | 2007-12-19 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insight on the mechanism of regulation of the MarR family of proteins: high-resolution crystal structure of a transcriptional repressor from Methanobacterium thermoautotrophicum.
J.Mol.Biol., 377, 2008
|
|
3SZT
 
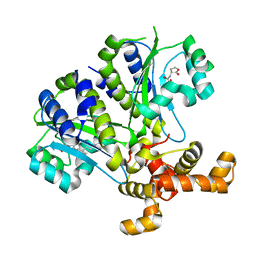 | | Quorum Sensing Control Repressor, QscR, Bound to N-3-oxo-dodecanoyl-L-Homoserine Lactone | | Descriptor: | N-3-OXO-DODECANOYL-L-HOMOSERINE LACTONE, Quorum-sensing control repressor, SODIUM ION | | Authors: | Churchill, M.E.A, Lintz, M.J. | | Deposit date: | 2011-07-19 | | Release date: | 2011-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of QscR, a Pseudomonas aeruginosa quorum sensing signal receptor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4MH8
 
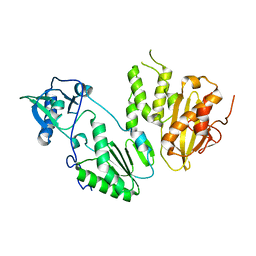 | |
6KTO
 
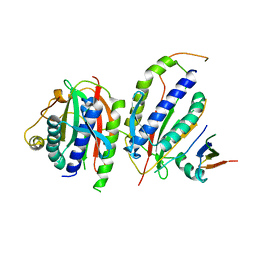 | | Crystal structure of human SHLD3-C-REV7-O-REV7-SHLD2 complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, Shieldin complex subunit 2, Shieldin complex subunit 3 | | Authors: | Liang, L, Yin, Y. | | Deposit date: | 2019-08-28 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4497633 Å) | | Cite: | Molecular basis for assembly of the shieldin complex and its implications for NHEJ.
Nat Commun, 11, 2020
|
|
6DSZ
 
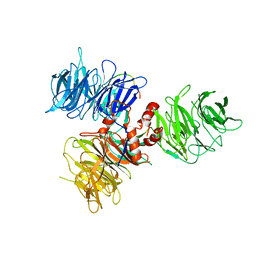 | |
1MNV
 
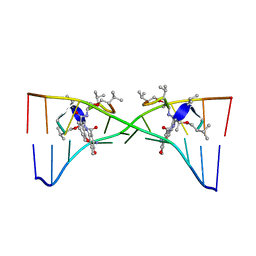 | | Actinomycin D binding to ATGCTGCAT | | Descriptor: | 5'-D(*AP*TP*GP*CP*TP*GP*CP*AP*T)-3', ACTINOMYCIN D | | Authors: | Hou, M.-H, Robinson, H, Gao, Y.-G, Wang, A.H.-J. | | Deposit date: | 2002-09-06 | | Release date: | 2002-11-22 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Actinomycin D Bound to the Ctg Triplet Repeat Sequences Linked to Neurological Diseases
Nucleic Acids Res., 30, 2002
|
|
4WWX
 
 | | Crystal structure of the core RAG1/2 recombinase | | Descriptor: | V(D)J recombination-activating protein 1, V(D)J recombination-activating protein 2, ZINC ION | | Authors: | Kim, M.S, Lapkouski, M, Yang, W, Gellert, M. | | Deposit date: | 2014-11-12 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2001 Å) | | Cite: | Crystal structure of the V(D)J recombinase RAG1-RAG2.
Nature, 518, 2015
|
|
3C1C
 
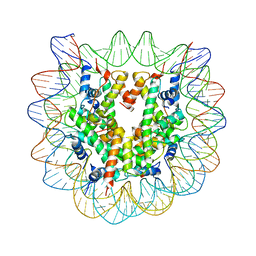 | | The effect of H3 K79 dimethylation and H4 K20 trimethylation on nucleosome and chromatin structure | | Descriptor: | Histone 2, H2bf, Histone H2A type 1, ... | | Authors: | Lu, X, Simon, M, Chodaparambil, J, Hansen, J, Shokat, K, Luger, K. | | Deposit date: | 2008-01-22 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2FUF
 
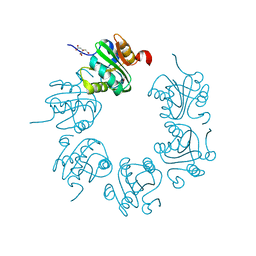 | |
6UT6
 
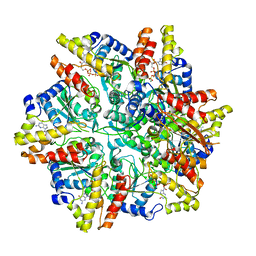 | | Cryo-EM structure of the Escherichia coli McrBC complex | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, 5-methylcytosine-specific restriction enzyme B, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Niu, Y, Suzuki, H, Hosford, C.J, Chappie, J.S, Walz, T. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural asymmetry governs the assembly and GTPase activity of McrBC restriction complexes.
Nat Commun, 11, 2020
|
|
7O5E
 
 | |
6ZS9
 
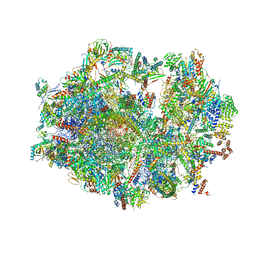 | | Human mitochondrial ribosome in complex with ribosome recycling factor | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
7P8K
 
 | | Crystal structure of in planta processed AvrRps4 in complex with the WRKY domain of RRS1 | | Descriptor: | Avirulence protein,Avirulence protein, Disease resistance protein RRS1, ZINC ION | | Authors: | Mukhi, N, Brown, H, Gorenkin, D, Ding, P, Bentham, A.R, Jones, J.D.G, Banfield, M.J. | | Deposit date: | 2021-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Perception of structurally distinct effectors by the integrated WRKY domain of a plant immune receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7A5I
 
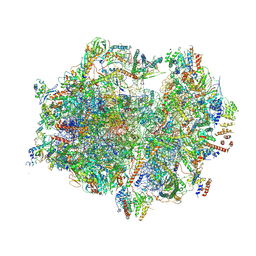 | | Structure of the human mitoribosome with A- P-and E-site mt-tRNAs | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
5EFY
 
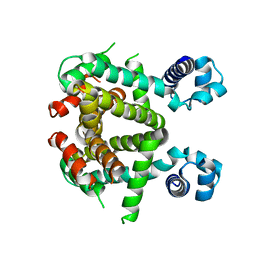 | | Apo-form of SCO3201 | | Descriptor: | Putative tetR-family transcriptional regulator | | Authors: | Waack, P, Hinrichs, W. | | Deposit date: | 2015-10-26 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of free and ligand-bound forms of the TetR/AcrR-like regulator SCO3201 from Streptomyces coelicolor suggest a novel allosteric mechanism.
Febs J., 2022
|
|
5ERI
 
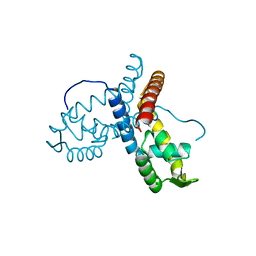 | |
1MK6
 
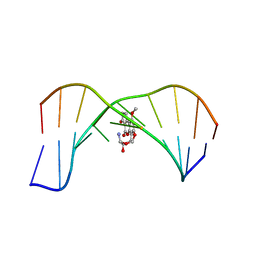 | | SOLUTION STRUCTURE OF THE 8,9-DIHYDRO-8-(N7-GUANYL)-9-HYDROXY-AFLATOXIN B1 ADDUCT MISPAIRED WITH DEOXYADENOSINE | | Descriptor: | 5'-D(*AP*CP*AP*TP*CP*GP*AP*TP*CP*T)-3', 5'-D(*AP*GP*AP*TP*AP*GP*AP*TP*GP*T)-3', 8,9-DIHYDRO-9-HYDROXY-AFLATOXIN B1 | | Authors: | Giri, I, Johnston, D.S, Stone, M.P. | | Deposit date: | 2002-08-28 | | Release date: | 2002-10-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | MISPAIRING OF THE 8,9-DIHYDRO-8-(N7-GUANYL)-9-HYDROXY-AFLATOXIN B1 ADDUCT WITH DEOXYADENOSINE RESULTS IN EXTRUSION OF THE MISMATCHED DA TOWARD THE MAJOR GROOVE
Biochemistry, 41, 2002
|
|
6X6T
 
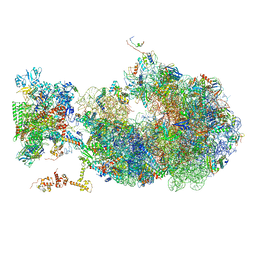 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B1 (TTC-B1) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6X9Q
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 27 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-06-03 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6X7F
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B2 (TTC-B2) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6X7K
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 24 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-05-30 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XDQ
 
 | | Cryo-EM structure of an Escherichia coli coupled transcription-translation complex B3 (TTC-B3) containing an mRNA with a 30 nt long spacer, transcription factors NusA and NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Ebright, R.H, Wang, C, Su, M. | | Deposit date: | 2020-06-11 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
