7KGE
 
 | |
7KGI
 
 | |
7KGF
 
 | |
7KGH
 
 | |
7KGD
 
 | |
7KGG
 
 | |
1HUQ
 
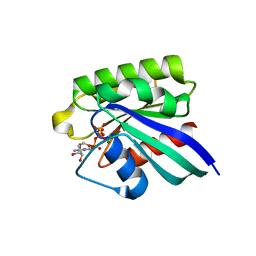 | | 1.8A CRYSTAL STRUCTURE OF THE MONOMERIC GTPASE RAB5C (MOUSE) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RAB5C | | Authors: | Merithew, E, Hatherly, S, Dumas, J.J, Lawe, D.C, Heller-Harrison, R, Lambright, D.G. | | Deposit date: | 2001-01-04 | | Release date: | 2001-02-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural plasticity of an invariant hydrophobic triad in the switch regions of Rab GTPases is a determinant of effector recognition.
J.Biol.Chem., 276, 2001
|
|
7KFU
 
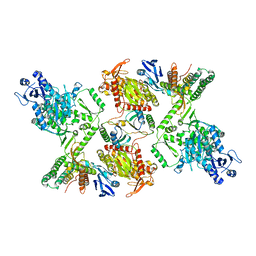 | | Cas6-RT-Cas1--Cas2 complex | | Descriptor: | Cas2, Cas6-RT-Cas1 | | Authors: | Hoel, C.M, Wang, J.Y, Doudna, J.A, Brohawn, S.G. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural coordination between active sites of a CRISPR reverse transcriptase-integrase complex.
Nat Commun, 12, 2021
|
|
6BRG
 
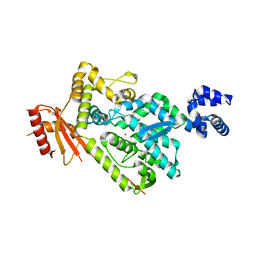 | | The SAM domain of mouse SAMHD1 is critical for its activation and regulation | | Descriptor: | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, MAGNESIUM ION | | Authors: | Buzovetsky, O, Tang, C, Knecht, K.M, Antonucci, J.M, Wu, L, Ji, X, Xiong, Y. | | Deposit date: | 2017-11-30 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The SAM domain of mouse SAMHD1 is critical for its activation and regulation.
Nat Commun, 9, 2018
|
|
7KFT
 
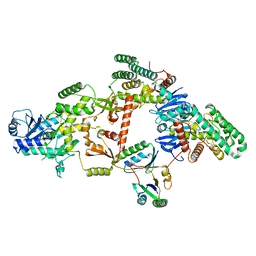 | | Partial Cas6-RT-Cas1--Cas2 complex | | Descriptor: | Cas2, Cas6-RT-Cas1 | | Authors: | Hoel, C.M, Wang, J.Y, Doudna, J.A, Brohawn, S.G. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural coordination between active sites of a CRISPR reverse transcriptase-integrase complex.
Nat Commun, 12, 2021
|
|
6D0H
 
 | |
1LB8
 
 | | Crystal structure of the Non-desensitizing GluR2 ligand binding core mutant (S1S2J-L483Y) in complex with AMPA at 2.3 resolution | | Descriptor: | (S)-ALPHA-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID, Glutamate receptor 2 | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
6D0Q
 
 | | Structure of a DNA retention-prone PCNA variant | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Kelch, B.A, Gaubitz, C. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.80051017 Å) | | Cite: | Effective mismatch repair depends on timely control of PCNA retention on DNA by the Elg1 complex.
Nucleic Acids Res., 47, 2019
|
|
7LGP
 
 | | DapE enzyme from Shigella flexneri | | Descriptor: | CHLORIDE ION, SODIUM ION, Succinyl-diaminopimelate desuccinylase, ... | | Authors: | Osipiuk, J, Endres, M, Becker, D.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | DapE enzyme from Shigella flexneri
To Be Published
|
|
6D0I
 
 | | ParT: Prs ADP-ribosylating toxin bound to cognate antitoxin ParS. L48M ParT, SeMet-substituted complex. | | Descriptor: | GLYCEROL, ParS: COG5642 (DUF2384) antitoxin fragment, ParT: COG5654 (RES domain) toxin | | Authors: | Piscotta, F.J, Jeffrey, P.D, Link, A.J. | | Deposit date: | 2018-04-10 | | Release date: | 2019-01-09 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | ParST is a widespread toxin-antitoxin module that targets nucleotide metabolism.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6D0R
 
 | | Structure of a DNA retention-prone PCNA variant | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Kelch, B.A, Gaubitz, C. | | Deposit date: | 2018-04-10 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.85856962 Å) | | Cite: | Effective mismatch repair depends on timely control of PCNA retention on DNA by the Elg1 complex.
Nucleic Acids Res., 47, 2019
|
|
4RNW
 
 | | Truncated version of the G303 Circular Permutation of Old Yellow Enzyme | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
5V5V
 
 | | Complex of NLGN2 with MDGA1 Ig1-Ig2 | | Descriptor: | MAM domain-containing glycosylphosphatidylinositol anchor protein 1, Neuroligin-2 | | Authors: | Gangwar, S.P, Machius, M, Rudenko, G. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.11 Å) | | Cite: | Molecular Mechanism of MDGA1: Regulation of Neuroligin 2:Neurexin Trans-synaptic Bridges.
Neuron, 94, 2017
|
|
1LB9
 
 | | Crystal structure of the Non-desensitizing GluR2 ligand binding core mutant (S1S2J-L483Y) in complex with antagonist DNQX at 2.3 A resolution | | Descriptor: | 6,7-DINITROQUINOXALINE-2,3-DIONE, Glutamate receptor 2, SULFATE ION | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
5V5W
 
 | |
4GWF
 
 | | Crystal structure of the tyrosine phosphatase SHP-2 with Y279C mutation | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qiu, W, Lin, A, Hutchinson, A, Romanov, V, Ruzanov, M, Thompson, C, Lam, K, Kisselman, G, Battaile, K, Chirgadze, N.Y. | | Deposit date: | 2012-09-03 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the tyrosine phosphatase SHP-2 with Y279C mutation
TO BE PUBLISHED
|
|
2AEM
 
 | | Crystal Structures of the MthK RCK Domain | | Descriptor: | Calcium-gated potassium channel mthK | | Authors: | Dong, J, Shi, N, Berke, I, Chen, L, Jiang, Y. | | Deposit date: | 2005-07-22 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the MthK RCK Domain and the Effect of Ca2+ on Gating Ring Stability
J.Biol.Chem., 280, 2005
|
|
5VC8
 
 | | Crystal structure of the WHSC1 PWWP1 domain | | Descriptor: | DNA (5'-D(P*CP*TP*(DN))-3'), Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION, ... | | Authors: | Qin, S, Tempel, W, Dong, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-28 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Histone and DNA binding ability studies of the NSD subfamily of PWWP domains.
Biochem.Biophys.Res.Commun., 569, 2021
|
|
1LBB
 
 | | Crystal structure of the GluR2 ligand binding domain mutant (S1S2J-N754D) in complex with kainate at 2.1 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamine receptor 2 | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-06-05 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
1LBC
 
 | | Crystal structure of GluR2 ligand binding core (S1S2J-N775S) in complex with cyclothiazide (CTZ) as well as glutamate at 1.8 A resolution | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamine Receptor 2, ... | | Authors: | Sun, Y, Olson, R, Horning, M, Armstrong, N, Mayer, M, Gouaux, E. | | Deposit date: | 2002-04-02 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of glutamate receptor desensitization.
Nature, 417, 2002
|
|
