6EKC
 
 | | Crystal structure of the BSD2 homolog of Arabidopsis thaliana bound to the octameric assembly of RbcL from Thermosynechococcus elongatus | | Descriptor: | DnaJ/Hsp40 cysteine-rich domain superfamily protein, Ribulose bisphosphate carboxylase large chain, ZINC ION | | Authors: | Aigner, H, Wilson, R.H, Bracher, A, Calisse, L, Bhat, J.Y, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2017-09-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2.
Science, 358, 2017
|
|
8AH8
 
 | |
2RRB
 
 | | Refinement of RNA binding domain in human Tra2 beta protein | | Descriptor: | cDNA FLJ40872 fis, clone TUTER2000283, highly similar to Homo sapiens transformer-2-beta (SFRS10) gene | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Sugano, S, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-06-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the dual RNA-recognition modes of human Tra2-beta RRM.
Nucleic Acids Res., 39, 2011
|
|
1TJL
 
 | | Crystal structure of transcription factor DksA from E. coli | | Descriptor: | DnaK suppressor protein, ZINC ION | | Authors: | Perederina, A, Svetlov, V, Vassylyeva, M.N, Artsimovitch, I, Yokoyama, S, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-06-06 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Regulation through the secondary channel--structural framework for ppGpp-DksA synergism during transcription
Cell(Cambridge,Mass.), 118, 2004
|
|
6ZTJ
 
 | | E. coli 70S-RNAP expressome complex in NusG-coupled state (38 nt intervening mRNA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTL
 
 | | E. coli 70S-RNAP expressome complex in collided state bound to NusG | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTO
 
 | | E. coli 70S-RNAP expressome complex in uncoupled state 1 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZU1
 
 | | E. coli 70S-RNAP expressome complex in uncoupled state 2 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
8JCH
 
 | |
5WMF
 
 | |
6O9M
 
 | | Structure of the human apo TFIIH | | Descriptor: | CDK-activating kinase assembly factor MAT1, General transcription factor IIH subunit 1, General transcription factor IIH subunit 2, ... | | Authors: | Yan, C.L, Dodd, T, He, Y, Tainer, J.A, Tsutakawa, S.E, Ivanov, I. | | Deposit date: | 2019-03-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Transcription preinitiation complex structure and dynamics provide insight into genetic diseases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4V14
 
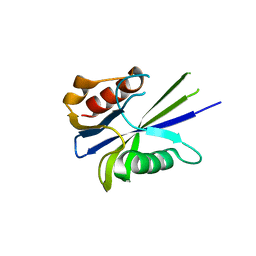 | |
3SZP
 
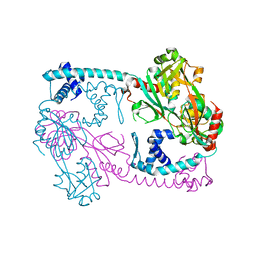 | | Full-length structure of the Vibrio cholerae virulence activator, AphB, a member of the LTTR protein family | | Descriptor: | Transcriptional regulator, LysR family | | Authors: | Taylor, J.L, De Silva, R.S, Kovacikova, G, Lin, W, Taylor, R.K, Skorupski, K, Kull, F.J. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | The crystal structure of AphB, a virulence gene activator from Vibrio cholerae, reveals residues that influence its response to oxygen and pH.
Mol.Microbiol., 83, 2012
|
|
2R1J
 
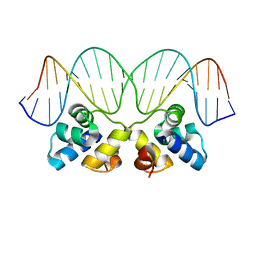 | | Crystal Structure of the P22 c2 Repressor protein in complex with the synthetic operator 9T | | Descriptor: | 5'-D(*DCP*DAP*DTP*DTP*DTP*DAP*DAP*DGP*DAP*DTP*DAP*DTP*DCP*DTP*DTP*DAP*DAP*DAP*DTP*DA)-3', 5'-D(*DTP*DAP*DTP*DTP*DTP*DAP*DAP*DGP*DAP*DTP*DAP*DTP*DCP*DTP*DTP*DAP*DAP*DAP*DTP*DG)-3', Repressor protein C2 | | Authors: | Williams, L.D, Koudelka, G.B, Watkins, D, Hsiao, C, Woods, K. | | Deposit date: | 2007-08-22 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | P22 c2 repressor-operator complex: mechanisms of direct and indirect readout
Biochemistry, 47, 2008
|
|
3T1B
 
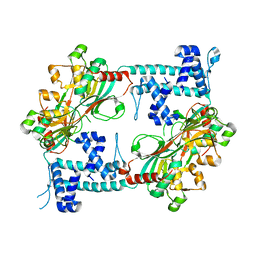 | | Crystal structure of the full-length AphB N100E variant | | Descriptor: | Transcriptional regulator, LysR family | | Authors: | Taylor, J.L, De Silva, R.S, Kovacikova, G, Lin, W, Taylor, R.K, Skorupski, K, Kull, F.J. | | Deposit date: | 2011-07-21 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of AphB, a virulence gene activator from Vibrio cholerae, reveals residues that influence its response to oxygen and pH.
Mol.Microbiol., 83, 2012
|
|
1W60
 
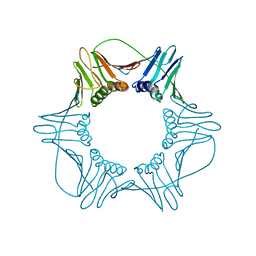 | | NATIVE HUMAN PCNA | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN | | Authors: | Kontopidis, G, Wu, S, Zheleva, D, Taylor, P, Mcinnes, C, Lane, D, Fischer, P, Walkinshaw, M. | | Deposit date: | 2004-08-11 | | Release date: | 2005-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural and Biochemical Studies of Human Proliferating Cell Nuclear Antigen Complexes Provide a Rationale for Cyclin Association and Inhibitor Design
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5LOE
 
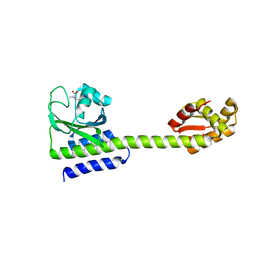 | | Structure of full length Cody from Bacillus subtilis in complex with Ile | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, ISOLEUCINE | | Authors: | Wilkinson, A.J, Levdikov, V.M, Blagova, E.V. | | Deposit date: | 2016-08-09 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Branched-chain Amino Acid and GTP-sensing Global Regulator, CodY, from Bacillus subtilis.
J. Biol. Chem., 292, 2017
|
|
5LOJ
 
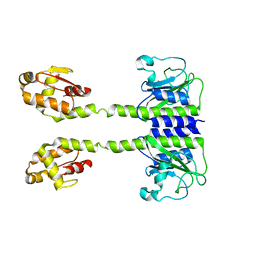 | |
5LNH
 
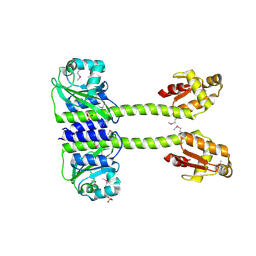 | | Structure of full length Unliganded CodY from Bacillus subtilis | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, SULFATE ION | | Authors: | Wilkinson, A.J, Levdikov, V.M, Blagova, E.V. | | Deposit date: | 2016-08-04 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Branched-chain Amino Acid and GTP-sensing Global Regulator, CodY, from Bacillus subtilis.
J. Biol. Chem., 292, 2017
|
|
8JHO
 
 | |
4E1R
 
 | |
4WYV
 
 | |
1MD0
 
 | |
1MJM
 
 | |
1MJO
 
 | |
