6NBT
 
 | |
8PDI
 
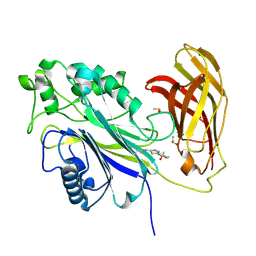 | | The phosphatase and C2 domains of SHIP1 with covalent Z1763271112 | | Descriptor: | (5-phenyl-1,3,4-thiadiazol-2-yl)methanimine, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Moreira, T, Pascoa, T.C, Bountra, C, Chalk, R, von Delft, F, Brennan, P.E, Gileadi, O. | | Deposit date: | 2023-06-12 | | Release date: | 2023-07-26 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 32, 2024
|
|
7ZGN
 
 | |
7WGP
 
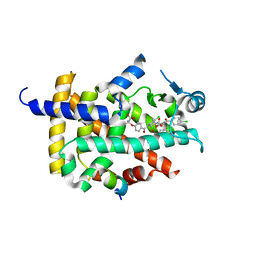 | | X-ray structure of human PPAR gamma ligand binding domain-fenofibric acid co-crystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Isoform 1 of Peroxisome proliferator-activated receptor gamma | | Authors: | Kamata, S, Honda, A, Akahane, M, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
7ZML
 
 | | Crystal structure of human RECQL5 helicase APO form in complex with engineered nanobody (Gluebody) G1-001 | | Descriptor: | ATP-dependent DNA helicase Q5, Gluebody G1-001, SULFATE ION, ... | | Authors: | Ye, M, Makola, M, Newman, J.A, Fairhead, M, MacLean, E, Krojer, T, Aitkenhead, H, Bountra, C, Gileadi, O, von Delft, F. | | Deposit date: | 2022-04-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Gluebodies improve crystal reliability and diversity through transferable nanobody mutations that introduce constitutive crystal contacts
To Be Published
|
|
7ZLU
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with UDP-2-deoxy-2-fluoro-D-glucose | | Descriptor: | 1,3-PROPANDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Roversi, P, Zitzmann, N, Bayo, Y, Ibba, R. | | Deposit date: | 2022-04-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint.
Iscience, 26, 2023
|
|
5FNM
 
 | | Native state mass spectrometry, surface plasmon resonance and X-ray crystallography correlate strongly as a fragment screening combination | | Descriptor: | (~{E})-3-(4-methoxyphenyl)but-2-enoic acid, CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Woods, L.A, Dolezal, O, Ren, B, Ryan, J.H, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2015-11-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance and X-Ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
6NC4
 
 | | FtsY-NG high-resolution | | Descriptor: | ACETATE ION, AMMONIUM ION, GLYCEROL, ... | | Authors: | Ataide, S.F, Faoro, C. | | Deposit date: | 2018-12-10 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the G-loop dynamics of E. coli FtsY NG domain.
J.Struct.Biol., 208, 2019
|
|
8PEA
 
 | |
6NCH
 
 | | Crystal structure of CDP-Chase: Raster data collection | | Descriptor: | D-ribose, PHOSPHATE ION, Phosphohydrolase (MutT/nudix family protein), ... | | Authors: | Miller, M.S, Shi, W, Gabelli, S.B. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
8W4D
 
 | |
7O4U
 
 | |
8PEB
 
 | |
5FIW
 
 | |
7WGO
 
 | | X-ray structure of human PPAR gamma ligand binding domain-bezafibrate co-rystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[P-[2-P-CHLOROBENZAMIDO)ETHYL]PHENOXY]-2-METHYLPROPIONIC ACID, Isoform 1 of Peroxisome proliferator-activated receptor gamma | | Authors: | Kamata, S, Honda, A, Akahane, M, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
7OCZ
 
 | |
7O6N
 
 | | Crystal structure of C. elegans ERH-2 PID-3 complex | | Descriptor: | Enhancer of rudimentary homolog 2, FORMIC ACID, Protein pid-3 | | Authors: | Falk, S, Ketting, R.F. | | Deposit date: | 2021-04-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis of PETISCO complex assembly during piRNA biogenesis in C. elegans .
Genes Dev., 35, 2021
|
|
8SKB
 
 | | Crystal Structure of GDP-mannose 3,5 epimerase de Myrciaria dubia in complex with NAD | | Descriptor: | GDP-mannose 3,5 epimerase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2023-04-19 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit camu-camu.
J.Exp.Bot., 75, 2024
|
|
5FJY
 
 | | Crystal structure of mouse kinesin light chain 2 (residues 161-480) | | Descriptor: | KINESIN LIGHT CHAIN 2, UNKNOWN PEPTIDE | | Authors: | Pernigo, S, Yip, Y.Y, Sanger, A, Xu, M, Dodding, M.P, Steiner, R.A. | | Deposit date: | 2015-10-14 | | Release date: | 2016-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The Light Chains of Kinesin-1 are Autoinhibited.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7WGN
 
 | | X-ray structure of human PPAR delta ligand binding domain-pemafibrate co-crystals obtained by co-crystallization | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, Peroxisome proliferator-activated receptor delta, octyl beta-D-glucopyranoside | | Authors: | Kamata, S, Honda, A, Akahane, M, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
5D5U
 
 | | Crystal structure of human Hsf1 with HSE DNA | | Descriptor: | Heat shock Element DNA, Heat shock factor protein 1 | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
7ZHI
 
 | |
7OCX
 
 | |
8SNZ
 
 | | X-ray Crystal Structure of FMN-bound long-chain flavodoxin from Rhodopseudomonas palustris | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Ansari, A, Khan, S.A, Miller, A.F. | | Deposit date: | 2023-04-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure, dynamics, and redox reactivity of an all-purpose flavodoxin.
J.Biol.Chem., 300, 2024
|
|
5VDO
 
 | | Human cyclic GMP-AMP synthase (cGAS) in complex with 2',2'-cGAMP | | Descriptor: | 2-amino-9-[(1R,3R,6R,8R,9R,11S,14R,16R,17R,18R)-16-(6-amino-9H-purin-9-yl)-3,11,17,18-tetrahydroxy-3,11-dioxido-2,4,7,10,12,15-hexaoxa-3,11-diphosphatricyclo[12.2.1.1~6,9~]octadec-8-yl]-1,9-dihydro-6H-purin-6-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Byrnes, L.J, Hall, J.D. | | Deposit date: | 2017-04-03 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.218 Å) | | Cite: | The catalytic mechanism of cyclic GMP-AMP synthase (cGAS) and implications for innate immunity and inhibition.
Protein Sci., 26, 2017
|
|
