1OOZ
 
 | | Deletion mutant of SUCCINYL-COA:3-KETOACID COA TRANSFERASE FROM PIG HEART | | Descriptor: | POTASSIUM ION, Succinyl-CoA:3-ketoacid-coenzyme A transferase | | Authors: | Coros, A.M, Swenson, L, Wolodko, W.T, Fraser, M.E. | | Deposit date: | 2003-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the CoA transferase from pig heart to 1.7 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3BHK
 
 | | Crystal structure of R49K mutant of monomeric sarcosine oxidase crystallized in phosphate as precipitant | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Hassan-Abdallah, A, Zhao, G, Chen, Z, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2007-11-28 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Arginine 49 is a bifunctional residue important in catalysis and biosynthesis of monomeric sarcosine oxidase: a context-sensitive model for the electrostatic impact of arginine to lysine mutations.
Biochemistry, 47, 2008
|
|
3F7N
 
 | | Crystal Structure of CheY triple mutant F14E, N59M, E89L complexed with BeF3- and Mn2+ | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY, GLYCEROL, ... | | Authors: | Pazy, Y, Collins, E.J, Bourret, R.B. | | Deposit date: | 2008-11-09 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Matching Biochemical Reaction Kinetics to the Timescales of Life: Structural Determinants That Influence the Autodephosphorylation Rate of Response Regulator Proteins.
J.Mol.Biol., 392, 2009
|
|
1OTB
 
 | | WILD TYPE PHOTOACTIVE YELLOW PROTEIN, P63 AT 295K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1P26
 
 | | Crystal structure of zinc(II)-d(GGCGCC)2 | | Descriptor: | 5'-D(*GP*GP*CP*GP*CP*C)-3', ZINC ION | | Authors: | Labiuk, S.L, Delbaere, L.T, Lee, J.S. | | Deposit date: | 2003-04-14 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Cobalt(II), nickel(II) and zinc(II) do not bind to intra-helical N(7)
guanine positions in the B-form crystal structure of d(GGCGCC)
J.Biol.Inorg.Chem., 8, 2003
|
|
3EYA
 
 | | Structural basis for membrane binding and catalytic activation of the peripheral membrane enzyme pyruvate oxidase from Escherichia coli | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Neumann, P, Weidner, A, Pech, A, Stubbs, M.T, Tittmann, K. | | Deposit date: | 2008-10-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for membrane binding and catalytic activation of the peripheral membrane enzyme pyruvate oxidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4O4P
 
 | | Structure of P450 BM3 A82F F87V in complex with S-omeprazol | | Descriptor: | 6-methoxy-2-{[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]sulfanyl}-1H-benzimidazole, Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leys, D. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Human P450-like oxidation of diverse proton pump inhibitor drugs by 'gatekeeper' mutants of flavocytochrome P450 BM3.
Biochem.J., 460, 2014
|
|
1Q6O
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-gulonaet 6-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-GULURONIC ACID 6-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-08-13 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.202 Å) | | Cite: | Structural Evidence for a 1,2-Enediolate Intermediate in the Reaction Catalyzed by 3-Keto-l-Gulonate 6-Phosphate Decarboxylase, a Member of the Orotidine 5'-Monophosphate Decarboxylase Suprafamily
Biochemistry, 42, 2003
|
|
1Q82
 
 | | Crystal Structure of CC-Puromycin bound to the A-site of the 50S ribosomal subunit | | Descriptor: | 23S ribosomal rna, 50S ribosomal protein L13P, 50S ribosomal protein L14P, ... | | Authors: | Hansen, J.L, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-20 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural Insights Into Peptide Bond Formation
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1QG3
 
 | |
3GZ1
 
 | | Crystal structure of IpgC in complex with the chaperone binding region of IpaB | | Descriptor: | Chaperone protein ipgC, GLYCEROL, Invasin ipaB | | Authors: | Lunelli, M, Lokareddy, R.K, Zychlinsky, A, Kolbe, M. | | Deposit date: | 2009-04-06 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | IpaB-IpgC interaction defines binding motif for type III secretion translocator
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1QZ5
 
 | | Structure of rabbit actin in complex with kabiramide C | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Klenchin, V.A, Allingham, J.S, King, R, Tanaka, J, Marriott, G, Rayment, I. | | Deposit date: | 2003-09-15 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Trisoxazole macrolide toxins mimic the binding of actin-capping proteins to actin
Nat.Struct.Biol., 10, 2003
|
|
2XZF
 
 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN THE WILD-TYPE LACTOCOCCUS LACTIS FPG (MUTM) AND AN OXIDIZED PYRIMIDINE CONTAINING DNA AT 293K | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*VETP*TP*TP*TP*CP*TP*CP*GP)-3', 5'-D(*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*AP*GP*CP)-3', FORMAMIDOPYRIMIDINE-DNA GLYCOSYLASE, ... | | Authors: | LeBihan, Y.V, Izquierdo, M.A, Coste, F, Culard, F, Gehrke, T.H, Essalhi, K, Aller, P, Carrel, T, Castaing, B. | | Deposit date: | 2010-11-25 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | 5-Hydroxy-5-Methylhydantoin DNA Lesion, a Molecular Trap for DNA Glycosylases
Nucleic Acids Res., 39, 2011
|
|
4O2X
 
 | | Structure of a malarial protein | | Descriptor: | Maltose-binding periplasmic protein, ATP-dependent Clp protease adaptor protein ClpS containing protein chimeric construct | | Authors: | AhYoung, A.P, Koehl, A, Cascio, D, Egea, P.F. | | Deposit date: | 2013-12-17 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a putative ClpS N-end rule adaptor protein from the malaria pathogen Plasmodium falciparum.
Protein Sci., 25, 2016
|
|
4O5I
 
 | |
1OYX
 
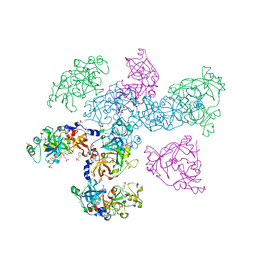 | | CRYSTAL STRUCTURE OF 3-MBT REPEATS OF LETHAL (3) MALIGNANT BRAIN TUMOR (SELENO-MET) AT 1.85 ANGSTROM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lethal(3)malignant brain tumor-like protein, SULFATE ION | | Authors: | Wang, W.K, Tereshko, V, Boccuni, P, MacGrogan, D, Nimer, S.D, Patel, D.J. | | Deposit date: | 2003-04-07 | | Release date: | 2003-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets.
Structure, 11, 2003
|
|
1OT6
 
 | |
2ZOK
 
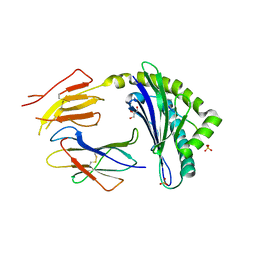 | | Crystal structure of H-2Db in complex with JHMV epitope S510 | | Descriptor: | 9-meric peptide from Spike glycoprotein, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Theodossis, A, Dunstone, M.A, Rossjohn, J. | | Deposit date: | 2008-05-22 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biological basis of CTL escape in coronavirus-infected mice.
J Immunol., 180, 2008
|
|
1P3J
 
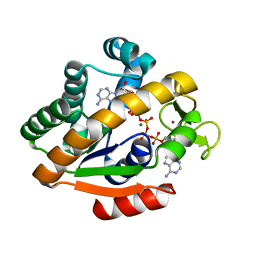 | | Adenylate Kinase from Bacillus subtilis | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bae, E, Phillips Jr, G.N. | | Deposit date: | 2003-04-17 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures and analysis of highly homologous psychrophilic, mesophilic, and thermophilic adenylate kinases.
J.Biol.Chem., 279, 2004
|
|
4OIP
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077, ATP, and CMPcPP | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
1P7C
 
 | | Crystal Structure of HSV1-TK complexed with TP5A | | Descriptor: | P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, SULFATE ION, THYMIDINE, ... | | Authors: | Gardberg, A, Shuvalova, L, Monnerjahn, C, Konrad, M, Lavie, A. | | Deposit date: | 2003-05-01 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the dual thymidine and thymidylate kinase activity of herpes thymidine kinases.
Structure, 11, 2003
|
|
1PA6
 
 | |
1OYN
 
 | | Crystal structure of PDE4D2 in complex with (R,S)-rolipram | | Descriptor: | ROLIPRAM, ZINC ION, cAMP-specific phosphodiesterase PDE4D2 | | Authors: | Huai, Q, Wang, H, Sun, Y, Kim, H.Y, Liu, Y, Ke, H. | | Deposit date: | 2003-04-05 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of PDE4D in complex with roliprams and implication on inhibitor selectivity
Structure, 11, 2003
|
|
3GPJ
 
 | | Crystal structure of the yeast 20S proteasome in complex with syringolin B | | Descriptor: | N-{[(1S)-2-methyl-1-{[(5S,8S)-5-(1-methylethyl)-2,7-dioxo-1,6-diazacyclododec-3-en-8-yl]carbamoyl}propyl]carbamoyl}-L-valine, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Huber, R, Kaiser, M. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic and structural studies on syringolin A and B reveal critical determinants of selectivity and potency of proteasome inhibition
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4OST
 
 | | Crystal structure of the S505C mutant of TAL effector dHax3 | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3 | | Authors: | Deng, D, Wu, J.P, Yan, C.Y, Pan, X.J, Yan, N. | | Deposit date: | 2014-02-13 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Revisiting the TALE repeat
Protein Cell, 5, 2014
|
|
