1R46
 
 | | Structure of human alpha-galactosidase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Garman, S.C, Garboczi, D.N. | | Deposit date: | 2003-10-03 | | Release date: | 2004-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The molecular defect leading to Fabry disease: structure of human alpha-galactosidase
J.Mol.Biol., 337, 2004
|
|
2AEC
 
 | | Crystal Structure of Human M340H-Beta1,4-Galactosyltransferase-I (M340H-B4GAL-T1) in Complex with GlcNAc-beta1,2-Man-alpha1,6-Man-beta-OR | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose, ... | | Authors: | Ramasamy, V, Ramakrishnan, B, Boeggeman, E, Ratner, D.M, Seeberger, P.H, Qasba, P.K. | | Deposit date: | 2005-07-22 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide Preferences of beta1,4-Galactosyltransferase-I: Crystal Structures of Met340His Mutant of Human beta1,4-Galactosyltransferase-I with a Pentasaccharide and Trisaccharides of the N-Glycan Moiety
J.Mol.Biol., 353, 2005
|
|
2KPL
 
 | | MAGI-1 PDZ1 / E6CT | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1, Protein E6 | | Authors: | Charbonnier, S, Nomine, Y, Ramirez, J, Luck, K, Stote, R.H, Trave, G, Kieffer, B, Atkinson, R.A. | | Deposit date: | 2009-10-16 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structural and dynamic response of MAGI-1 PDZ1 with non-canonical domain boundaries to binding of human papillomavirus (HPV) E6
J.Mol.Biol., 2011
|
|
1R47
 
 | | Structure of human alpha-galactosidase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, ... | | Authors: | Garman, S.C, Garboczi, D.N. | | Deposit date: | 2003-10-03 | | Release date: | 2004-03-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The molecular defect leading to Fabry disease: structure of human alpha-galactosidase
J.Mol.Biol., 337, 2004
|
|
2H7C
 
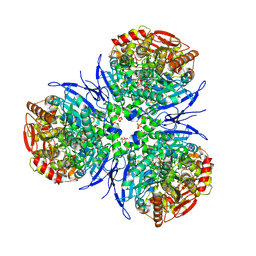 | | Crystal structure of human carboxylesterase in complex with Coenzyme A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COENZYME A, ... | | Authors: | Bencharit, S, Edwards, C.C, Morton, C.L, Howard-Williams, E.L, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2006-06-02 | | Release date: | 2006-08-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multisite promiscuity in the processing of endogenous substrates by human carboxylesterase 1
J.Mol.Biol., 363, 2006
|
|
2FQQ
 
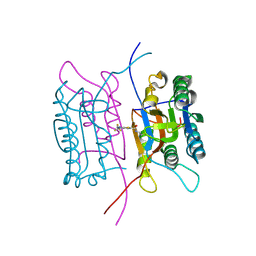 | | Crystal structure of human caspase-1 (Cys285->Ala, Cys362->Ala, Cys364->Ala, Cys397->Ala) in complex with 1-methyl-3-trifluoromethyl-1H-thieno[2,3-c]pyrazole-5-carboxylic acid (2-mercapto-ethyl)-amide | | Descriptor: | 1-METHYL-3-TRIFLUOROMETHYL-1H-THIENO[2,3-C]PYRAZOLE-5-CARBOXYLIC ACID (2-MERCAPTO-ETHYL)-AMIDE, Caspase-1 | | Authors: | Scheer, J.M, Wells, J.A, Romanowski, M.J. | | Deposit date: | 2006-01-18 | | Release date: | 2006-05-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A common allosteric site and mechanism in caspases
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2G07
 
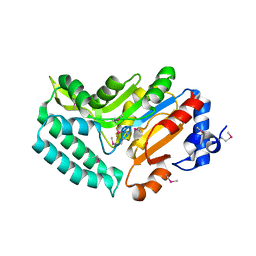 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, phospho-enzyme intermediate analog with Beryllium fluoride | | Descriptor: | Cytosolic 5'-nucleotidase III, MAGNESIUM ION | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
2KW6
 
 | | Solution NMR Structure of Cyclin-dependent kinase 2-associated protein 1 (CDK2-associated protein 1; oral cancer suppressor Deleted in oral cancer 1, DOC-1) from H.sapiens, Northeast Structural Genomics Consortium Target Target HR3057H | | Descriptor: | Cyclin-dependent kinase 2-associated protein 1 | | Authors: | Ertekin, A, Aramini, J.M, Rossi, P, Lee, A.B, Jiang, M, Ciccosanti, C.T, Xiao, R, Swapna, G.V.T, Rost, B, Everett, J.K, Acton, T.B, Prestegard, J.H, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Human cyclin-dependent kinase 2-associated protein 1 (CDK2AP1) is dimeric in its disulfide-reduced state, with natively disordered N-terminal region.
J.Biol.Chem., 287, 2012
|
|
2H4W
 
 | |
1S1T
 
 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, PHOSPHATE ION, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
2JJK
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE -1- PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH A DUAL BINDING AMP SITE INHIBITOR | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N,N'-(heptane-1,7-diyldicarbamoyl)bis(3-chlorobenzenesulfonamide) | | Authors: | Ruf, A, Joseph, C, Benz, J, Fol, B, Tetaz, T, Hebeisen, P. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric Fbpase Inhibitors Gain 10(5) Times in Potency When Simultaneously Binding Two Neighboring AMP Sites.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2G08
 
 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, product-transition complex analog with Aluminum fluoride | | Descriptor: | ALUMINUM FLUORIDE, Cytosolic 5'-nucleotidase III, MAGNESIUM ION | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
2LF4
 
 | |
2LCP
 
 | | NMR structure of calcium loaded, un-myristoylated human NCS-1 | | Descriptor: | CALCIUM ION, Neuronal calcium sensor 1 | | Authors: | Heidarsson, P.O, Bjerrum-Bohr, I.J, Bellucci, L, Gitte, J, Corni, S, Poulsen, F.M, Finn, B.E, Di Felice, R, Kragelund, B. | | Deposit date: | 2011-05-05 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-terminal tail of human neuronal calcium sensor 1 regulates the conformational stability of the ca(2+)-activated state.
J.Mol.Biol., 417, 2012
|
|
2L8J
 
 | | GABARAPL-1 NBR1-LIR complex structure | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, NBR1-LIR peptide | | Authors: | Rogov, V.V, Rozenknop, A, Rogova, N.Y, Loehr, F, Guentert, P, Dikic, I, Doetsch, V. | | Deposit date: | 2011-01-17 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of the Interaction of GABARAPL-1 with the LIR Motif of NBR1.
J.Mol.Biol., 410, 2011
|
|
2LGY
 
 | | Ubiquitin-like domain from HOIL-1 | | Descriptor: | RanBP-type and C3HC4-type zinc finger-containing protein 1 | | Authors: | Beasley, S.A, Shaw, G.S. | | Deposit date: | 2011-08-03 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the E3 ligase HOIL-1 Ubl domain.
Protein Sci., 21, 2012
|
|
2J0T
 
 | | Crystal Structure of the Catalytic Domain of MMP-1 in Complex with the Inhibitory Domain of TIMP-1 | | Descriptor: | CALCIUM ION, INTERSTITIAL COLLAGENASE, METALLOPROTEINASE INHIBITOR 1, ... | | Authors: | Iyer, S, Wei, S, Brew, K, Acharya, K.R. | | Deposit date: | 2006-08-04 | | Release date: | 2006-10-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Matrix Metalloproteinase-1 in Complex with the Inhibitory Domain of Tissue Inhibitor of Metalloproteinase-1.
J.Biol.Chem., 282, 2007
|
|
1HZH
 
 | | CRYSTAL STRUCTURE OF THE INTACT HUMAN IGG B12 WITH BROAD AND POTENT ACTIVITY AGAINST PRIMARY HIV-1 ISOLATES: A TEMPLATE FOR HIV VACCINE DESIGN | | Descriptor: | IMMUNOGLOBULIN HEAVY CHAIN, IMMUNOGLOBULIN LIGHT CHAIN,Uncharacterized protein, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saphire, E.O, Burton, D.R, Wilson, I.A. | | Deposit date: | 2001-01-24 | | Release date: | 2001-08-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design.
Science, 293, 2001
|
|
2J94
 
 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 5-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}-1H-1,2,4-TRIAZOLE-3-SULFONAMIDE, CALCIUM ION, COAGULATION FACTOR X | | Authors: | Chan, C, Borthwick, A.D, Brown, D, Campbell, M, Chaudry, L, Chung, C.W, Convery, M.A, Hamblin, J.N, Johnstone, L, Kelly, H.A, Kleanthous, S, Burns-Kurtis, C.L, Patikis, A, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Whitworth, C, Young, R.J, Zhou, P. | | Deposit date: | 2006-11-02 | | Release date: | 2007-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Factor Xa Inhibitors: S1 Binding Interactions of a Series of N-{(3S)-1-[(1S)-1-Methyl-2-Morpholin-4-Yl-2-Oxoethyl]-2-Oxopyrrolidin-3-Yl}Sulfonamides.
J.Med.Chem., 50, 2007
|
|
2J9H
 
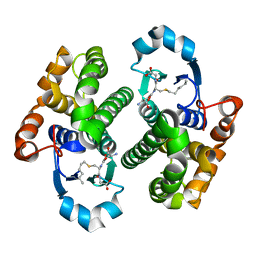 | | Crystal structure of human glutathione-S-transferase P1-1 cys-free mutant in complex with S-hexylglutathione at 2.4 A resolution | | Descriptor: | GLUTATHIONE S-TRANSFERASE P, S-HEXYLGLUTATHIONE | | Authors: | Tars, K, Hegazy, U.M, Hellman, U, Mannervik, B. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Modulating Catalytic Activity by Unnatural Amino Acid Residues in a Gsh-Binding Loop of Gst P1-1.
J.Mol.Biol., 376, 2008
|
|
1H4P
 
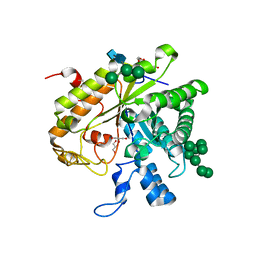 | | Crystal structure of exo-1,3-beta glucanse from Saccharomyces cerevisiae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUCAN 1,3-BETA-GLUCOSIDASE I/II, GLYCEROL, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2001-05-11 | | Release date: | 2003-10-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Er Protein Folding Sensor Udp-Glucose Glycoprotein:Glucosyltransferase Modifies Substrates Distant to Local Changes in Glycoprotein Conformation.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2ERM
 
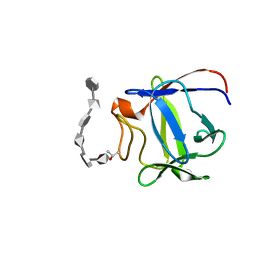 | | Solution structure of a biologically active human FGF-1 monomer, complexed to a hexasaccharide heparin-analogue | | Descriptor: | 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, ISOPROPYL ALCOHOL | | Authors: | Canales, A, Lozano, R, Nieto, P.M, Martin-Lomas, M, Gimenez-Gallego, G, Jimenez-Barbero, J. | | Deposit date: | 2005-10-25 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a human FGF-1 monomer, activated by a hexasaccharide heparin-analogue.
Febs J., 273, 2006
|
|
1E6X
 
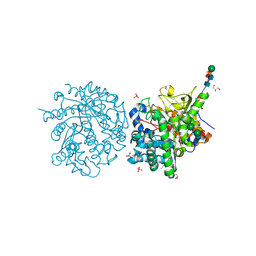 | | MYROSINASE FROM SINAPIS ALBA with a bound transition state analogue,D-glucono-1,5-lactone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-glucono-1,5-lactone, ... | | Authors: | Burmeister, W.P. | | Deposit date: | 2000-08-23 | | Release date: | 2001-01-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High Resolution X-Ray Crystallography Shows that Ascorbate is a Cofactor for Myrosinase and Substitutes for the Function of the Catalytic Base
J.Biol.Chem., 275, 2000
|
|
2AE7
 
 | | Crystal Structure of Human M340H-Beta1,4-Galactosyltransferase-I (M340H-B4GAL-T1) in Complex with Pentasaccharide | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, ... | | Authors: | Ramasamy, V, Ramakrishnan, B, Boeggeman, E, Ratner, D.M, Seeberger, P.H, Qasba, P.K. | | Deposit date: | 2005-07-21 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide Preferences of beta1,4-Galactosyltransferase-I: Crystal Structures of Met340His Mutant of Human beta1,4-Galactosyltransferase-I with a Pentasaccharide and Trisaccharides of the N-Glycan Moiety
J.Mol.Biol., 353, 2005
|
|
1GAH
 
 | | GLUCOAMYLASE-471 COMPLEXED WITH ACARBOSE | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLUCOAMYLASE-471, alpha-D-mannopyranose, ... | | Authors: | Aleshin, A.E, Stoffer, B, Firsov, L.M, Svensson, B, Honzatko, R.B. | | Deposit date: | 1996-03-06 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic complexes of glucoamylase with maltooligosaccharide analogs: relationship of stereochemical distortions at the nonreducing end to the catalytic mechanism.
Biochemistry, 35, 1996
|
|
