3VL9
 
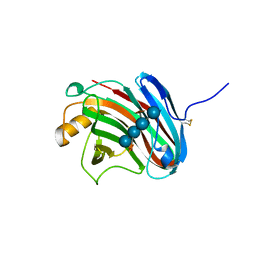 | | Crystal structure of xeg-xyloglucan | | Descriptor: | Xyloglucan-specific endo-beta-1,4-glucanase A, beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
1P6W
 
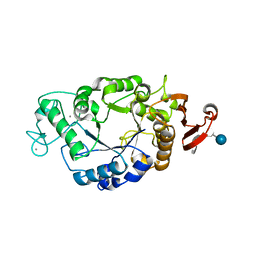 | | Crystal structure of barley alpha-amylase isozyme 1 (AMY1) in complex with the substrate analogue, methyl 4I,4II,4III-tri-thiomaltotetraoside (thio-DP4) | | Descriptor: | CALCIUM ION, PROTEIN (Alpha-amylase type A isozyme), alpha-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose, ... | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2003-04-30 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of barley alpha-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding: a pair of sugar tongs
Structure, 11, 2003
|
|
2EMT
 
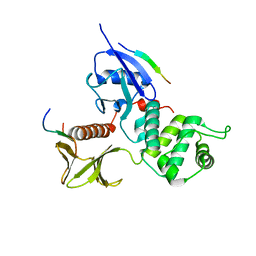 | | Crystal Structure Analysis of the radixin FERM domain complexed with adhesion molecule PSGL-1 | | Descriptor: | P-selectin glycoprotein ligand 1, Radixin | | Authors: | Takai, Y, Kitano, K, Terawaki, S, Maesaki, R, Hakoshima, T. | | Deposit date: | 2007-03-28 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of PSGL-1 binding to ERM proteins
Genes Cells, 12, 2007
|
|
2J51
 
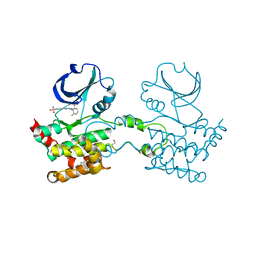 | | Crystal structure of Human STE20-like kinase bound to 5-Amino-3-((4-(aminosulfonyl)phenyl)amino) -N-(2,6-difluorophenyl)-1H-1,2,4-triazole- 1-carbothioamide | | Descriptor: | 1,2-ETHANEDIOL, 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, STE20-LIKE SERINE/THREONINE-PROTEIN KINASE, ... | | Authors: | Pike, A.C.W, Rellos, P, Fedorov, O, Keates, T, Salah, E, Savitsky, P, Papagrigoriou, E, Turnbull, A.P, Debreczeni, J.E, von Delft, F, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S. | | Deposit date: | 2006-09-08 | | Release date: | 2006-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activation Segment Dimerization: A Mechanism for Kinase Autophosphorylation of Non-Consensus Sites.
Embo J., 27, 2008
|
|
2NL9
 
 | | Crystal structure of the Mcl-1:Bim BH3 complex | | Descriptor: | Bcl-2-like protein 11, CHLORIDE ION, FUSION PROTEIN CONSISTING OF Induced myeloid leukemia cell differentiation protein Mcl-1 homolog, ... | | Authors: | Czabotar, P.E, Colman, P.M. | | Deposit date: | 2006-10-19 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the degradation of Mcl-1 induced by BH3 domains.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2NLP
 
 | |
1GZ1
 
 | | Mutant D416A of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with methyl-cellobiosyl-4-deoxy-4-thio-beta-D-cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE II, SODIUM ION, ... | | Authors: | Varrot, A, Frandsen, T, Driguez, H, Davies, G.J. | | Deposit date: | 2002-05-13 | | Release date: | 2003-02-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Humicola Insolens Cellobiohydrolase Cel6A D416A Mutant in Complex with a Non-Hydrolysable Substrate Analogue, Methyl Cellobiosyl-4-Thio-Beta-Cellobioside, at 1.9 A
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2JEO
 
 | | Crystal structure of human uridine-cytidine kinase 1 | | Descriptor: | URIDINE-CYTIDINE KINASE 1 | | Authors: | Kosinska, U, Stenmark, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E.P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Uppsten, M, Thorsell, A.G, van den Berg, S, Weigelt, J, Welin, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Human Uridine-Cytidine Kinase 1
To be Published
|
|
3LKW
 
 | | Crystal Structure of Dengue Virus 1 NS2B/NS3 protease active site mutant | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chandramouli, S, Joseph, J.S, Daudenarde, S, Gatchalian, J, Cornillez-Ty, C, Kuhn, P. | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Serotype-specific structural differences in the protease-cofactor complexes of the dengue virus family.
J.Virol., 84, 2010
|
|
2E0J
 
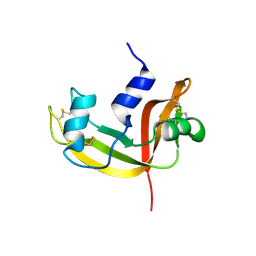 | | Mutant Human Ribonuclease 1 (R31L, R32L) | | Descriptor: | Ribonuclease | | Authors: | Yamada, H, Tamada, T, Kosaka, M, Kuroki, R. | | Deposit date: | 2006-10-10 | | Release date: | 2007-08-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 'Crystal lattice engineering,' an approach to engineer protein crystal contacts by creating intermolecular symmetry: crystallization and structure determination of a mutant human RNase 1 with a hydrophobic interface of leucines
Protein Sci., 16, 2007
|
|
4OVE
 
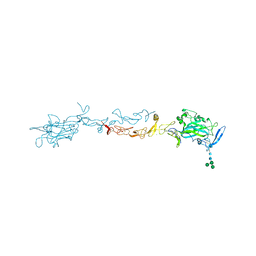 | | X-ray Crystal Structure of Mouse Netrin-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Meier, M, Nikodemus, D, Reuten, R, Patel, T.R, Orriss, G, Okun, N, Koch, M, Stetefeld, J. | | Deposit date: | 2014-02-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural Decoding of the Netrin-1/UNC5 Interaction and its Therapeutical Implications in Cancers.
Cancer Cell, 29, 2016
|
|
3USD
 
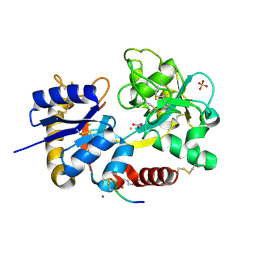 | | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Imidazol (1,2 a) pyridine3-yl-acitic acid at 2.4 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-terminal peptide of Lactotransferrin, ... | | Authors: | Shukla, P.K, Gautam, L, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Imidazol (1,2 a) pyridine3-yl-acitic acid at 2.4 A Resolution
To be Published
|
|
1Y2D
 
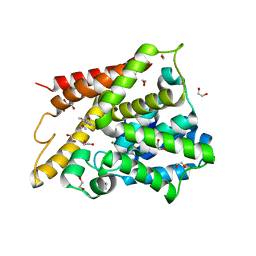 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With 1-(4-methoxy-phenyl)-3,5-dimethyl-1H-pyrazole-4-carboxylic acid ethyl ester | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-METHOXYPHENYL)-3,5-DIMETHYL-1H-PYRAZOLE-4-CARBOXYLIC ACID ETHYL ESTER, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Card, G.L, Blasdel, L, England, B.P, Zhang, C, Suzuki, Y, Gillette, S, Fong, D, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design
Nat.Biotechnol., 23, 2005
|
|
1BQS
 
 | | THE CRYSTAL STRUCTURE OF MUCOSAL ADDRESSIN CELL ADHESION MOLECULE-1 (MADCAM-1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (MUCOSAL ADDRESSIN CELL ADHESION MOLECULE-1) | | Authors: | Tan, K, Casasnovas, J.M, Liu, J.H, Briskin, M.J, Springer, T.A, Wang, J.-H. | | Deposit date: | 1998-08-18 | | Release date: | 1999-08-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of immunoglobulin superfamily domains 1 and 2 of MAdCAM-1 reveals novel features important for integrin recognition.
Structure, 6, 1998
|
|
2ERL
 
 | | PHEROMONE ER-1 FROM | | Descriptor: | ETHANOL, MATING PHEROMONE ER-1 | | Authors: | Anderson, D.H, Weiss, M.S, Eisenberg, D. | | Deposit date: | 1995-12-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A challenging case for protein crystal structure determination: the mating pheromone Er-1 from Euplotes raikovi.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2ERF
 
 | |
2JEV
 
 | | Crystal structure of human spermine,spermidine acetyltransferase in complex with a bisubstrate analog (N1-acetylspermine-S-CoA). | | Descriptor: | (3R)-27-AMINO-3-HYDROXY-2,2-DIMETHYL-4,8,14-TRIOXO-12-THIA-5,9,15,19,24-PENTAAZAHEPTACOS-1-YL [(2S,3R,4S,5S)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, DIAMINE ACETYLTRANSFERASE 1 | | Authors: | Hegde, S.S, Chandler, J, Vetting, M.W, Yu, M, Blanchard, J.S. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic and Structural Analysis of Human Spermidine/Spermine N(1)-Acetyltransferase.
Biochemistry, 46, 2007
|
|
3CVK
 
 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[1-(3,3-Dimethyl-butyl)-4-hydroxy-2-oxo-1,2,4a,5,6,7-hexahydro-pyrrolo[1,2-b]pyridazin-3-yl]-1,1-dioxo-1,2-dihydro -1lambda6-benzo[1,2,4]thiadiazin-7-yl}-methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2008-04-18 | | Release date: | 2009-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Hexahydro-pyrrolo- and hexahydro-1H-pyrido[1,2-b]pyridazin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2JMG
 
 | | Solution structure of V7R mutant of HIV-1 myristoylated matrix protein | | Descriptor: | Gag polyprotein, MYRISTIC ACID | | Authors: | Saad, J.S, Loeliger, E, Luncsford, P, Liriano, M, Tai, J, Kim, A, Miller, J, Joshi, A, Freed, E.O, Summers, M.F. | | Deposit date: | 2006-11-11 | | Release date: | 2007-02-06 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Point mutations in the HIV-1 matrix protein turn off the myristyl switch.
J.Mol.Biol., 366, 2007
|
|
1Z2N
 
 | | Inositol 1,3,4-trisphosphate 5/6-kinase complexed Mg2+/ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, inositol 1,3,4-trisphosphate 5/6-kinase | | Authors: | Miller, G.J, Wilson, M.P, Majerus, P.W, Hurley, J.H. | | Deposit date: | 2005-03-08 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Specificity determinants in inositol polyphosphate synthesis: crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase.
Mol.Cell, 18, 2005
|
|
2IS9
 
 | | Structure of yeast DCN-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Defective in cullin neddylation protein 1, ... | | Authors: | Yang, X, Zhou, J, Sun, L, Wei, Z, Gao, J, Gong, W, Xu, R.M, Rao, Z, Liu, Y. | | Deposit date: | 2006-10-16 | | Release date: | 2007-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for the function of DCN-1 in protein Neddylation.
J.Biol.Chem., 282, 2007
|
|
2IJ0
 
 | | Structural basis of T cell specificity and activation by the bacterial superantigen toxic shock syndrome toxin-1 | | Descriptor: | Toxic shock syndrome toxin-1, penultimate affinity-matured variant of hVbeta 2.1, D10 | | Authors: | Moza, B, Varma, A.K, Buonpane, R.A, Zhu, P, Herfst, C.A, Nicholson, M.J, Wilbuer, A.K, Nulifer, S, Wucherpfenning, K.W, McCormick, J.K, Kranz, D.M, Sundberg, E.J. | | Deposit date: | 2006-09-28 | | Release date: | 2007-02-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of T-cell specificity and activation by the bacterial superantigen TSST-1.
Embo J., 26, 2007
|
|
2C6L
 
 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | Descriptor: | 4-({5-[(4-AMINOCYCLOHEXYL)AMINO][1,2,4]TRIAZOLO[1,5-A]PYRIMIDIN-7-YL}AMINO)BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2005-11-10 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Triazolo[1,5-A]Pyrimidines as Novel Cdk2 Inhibitors: Protein Structure-Guided Design and Sar.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2OI6
 
 | | E. coli GlmU- Complex with UDP-GlcNAc, CoA and GlcN-1-PO4 | | Descriptor: | 2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, Bifunctional protein glmU, COBALT (II) ION, ... | | Authors: | Olsen, L.R, Vetting, M.W, Roderick, S.L. | | Deposit date: | 2007-01-10 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the E. coli bifunctional GlmU acetyltransferase active site with substrates and products.
Protein Sci., 16, 2007
|
|
2J4W
 
 | | Structure of a Plasmodium vivax apical membrane antigen 1-Fab F8.12.19 complex | | Descriptor: | APICAL MEMBRANE ANTIGEN 1, FAB FRAGMENT OF MONOCLONAL ANTIBODY F8.12.19 | | Authors: | Igonet, S, Vulliez-Le Normand, B, Faure, G, Riottot, M.M, Kocken, C.H.M, Thomas, A.W, Bentley, G.A. | | Deposit date: | 2006-09-07 | | Release date: | 2007-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cross-Reactivity Studies of an Anti-Plasmodium Vivax Apical Membrane Antigen 1 Monoclonal Antibody: Binding and Structural Characterisation.
J.Mol.Biol., 366, 2007
|
|
