8VJJ
 
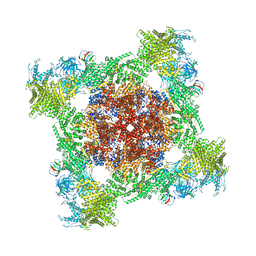 | | Structure of mouse RyR1 (EGTA-only dataset) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ... | | Authors: | Weninger, G, Marks, A.R. | | Deposit date: | 2024-01-07 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural insights into the regulation of RyR1 by S100A1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VK4
 
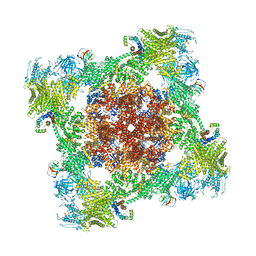 | |
8UW9
 
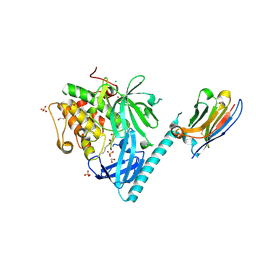 | | Structure of AKT1(E17K) with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-({4-[(2P)-2-(2-aminopyridin-3-yl)-5-phenyl-3H-imidazo[4,5-b]pyridin-3-yl]phenyl}methyl)-2-(2-fluoro-4-formyl-3-hydroxyphenyl)acetamide, ... | | Authors: | Craven, G.B, Taunton, J. | | Deposit date: | 2023-11-06 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutant-selective AKT1 inhibitors via lysine targeting and neo-zinc chelation
To Be Published
|
|
8UVY
 
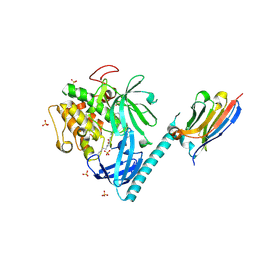 | | Structure of AKT1(E17K) with compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 4-{2-[({4-[(2P)-2-(2-aminopyridin-3-yl)-5-phenyl-3H-imidazo[4,5-b]pyridin-3-yl]phenyl}methyl)amino]ethyl}-2-hydroxybenzaldehyde, NB41, ... | | Authors: | Craven, G.B, Taunton, J. | | Deposit date: | 2023-11-05 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Mutant-selective AKT1 inhibitors via lysine targeting and neo-zinc chelation
To Be Published
|
|
8UW7
 
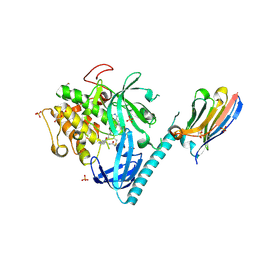 | | Structure of AKT1(WT) with compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 4-{2-[({4-[(2P)-2-(2-aminopyridin-3-yl)-5-phenyl-3H-imidazo[4,5-b]pyridin-3-yl]phenyl}methyl)amino]ethyl}-2-hydroxybenzaldehyde, NB41, ... | | Authors: | Craven, G.B, Taunton, J. | | Deposit date: | 2023-11-06 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Mutant-selective AKT1 inhibitors via lysine targeting and neo-zinc chelation
To Be Published
|
|
3COG
 
 | | Crystal structure of human cystathionase (Cystathionine gamma lyase) in complex with DL-propargylglycine | | Descriptor: | (2S)-2-aminopent-4-enoic acid, Cystathionine gamma-lyase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Collins, R, Karlberg, T, Lehtio, L, Arrowsmith, C.H, Berglund, H, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Schuler, H, Svensson, L, Thorsell, A.G, Tresaugues, L, Van den Berg, S, Sagermark, J, Busam, R.D, Welin, M, Weigelt, J, Wikstrom, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-28 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the inhibition mechanism of human cystathionine gamma-lyase, an enzyme responsible for the production of H(2)S.
J.Biol.Chem., 284, 2009
|
|
3EY6
 
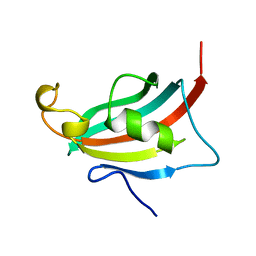 | | Crystal structure of the FK506-binding domain of human FKBP38 | | Descriptor: | FK506-binding protein 8 | | Authors: | Parthier, C, Maestre-Martinez, M, Neumann, P, Edlich, F, Fischer, G, Luecke, C, Stubbs, M.T. | | Deposit date: | 2008-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A charge-sensitive loop in the FKBP38 catalytic domain modulates Bcl-2 binding.
J.Mol.Recognit., 24, 2011
|
|
3ELP
 
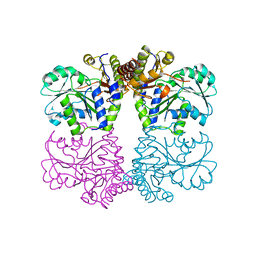 | | Structure of cystationine gamma lyase | | Descriptor: | Cystathionine gamma-lyase | | Authors: | Sun, Q, Sivaraman, J. | | Deposit date: | 2008-09-23 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the inhibition mechanism of human cystathionine gamma-lyase, an enzyme responsible for the production of H(2)S
J.Biol.Chem., 284, 2009
|
|
3G8H
 
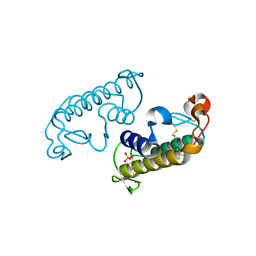 | |
3DFA
 
 | | Crystal structure of kinase domain of calcium-dependent protein kinase cgd3_920 from Cryptosporidium parvum | | Descriptor: | Calcium-dependent protein kinase cgd3_920 | | Authors: | Wernimont, A.K, Lew, J, Lin, Y.H, Hassanali, A, Khuu, C, Alam, Z, Kozieradzki, I, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Wilkstrom, M, Edwards, A.M, Hui, R, Artz, J.D, Xiao, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-11 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of kinase domain of calcium-dependent protein kinase cgd3_920 from Cryptosporidium parvum.
To be Published
|
|
3G8G
 
 | |
1TTX
 
 | | Solution Structure of human beta parvalbumin (oncomodulin) refined with a paramagnetism based strategy | | Descriptor: | CALCIUM ION, Oncomodulin | | Authors: | Babini, E, Bertini, I, Capozzi, F, Del Bianco, C, Hollender, D, Kiss, T, Luchinat, C, Quattrone, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-23 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human beta-Parvalbumin and Structural Comparison with Its Paralog alpha-Parvalbumin and with Their Rat Orthologs(,)
Biochemistry, 43, 2004
|
|
1YHP
 
 | | Solution Structure of Ca2+-free DdCAD-1 | | Descriptor: | Calcium-dependent cell adhesion molecule-1 | | Authors: | Lin, Z, Huang, H.B, Siu, C.H, Yang, D.W. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the adhesion molecule DdCAD-1 reveal new insights into Ca(2+)-dependent cell-cell adhesion
Nat.Struct.Mol.Biol., 13, 2006
|
|
1YMG
 
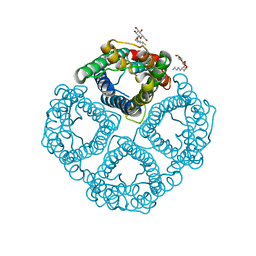 | | The Channel Architecture of Aquaporin O at 2.2 Angstrom Resolution | | Descriptor: | Lens fiber major intrinsic protein, nonyl beta-D-glucopyranoside | | Authors: | Harries, W.E.C, Akhavan, D, Miercke, L.J.W, Khademi, S, Stroud, R.M. | | Deposit date: | 2005-01-20 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Channel Architecture of Aquaporin 0 at a 2.2-A Resolution
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1XZZ
 
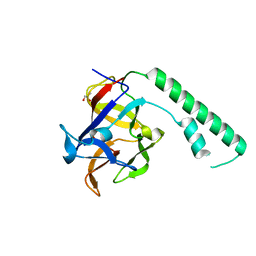 | | Crystal structure of the ligand binding suppressor domain of type 1 inositol 1,4,5-trisphosphate receptor | | Descriptor: | GLYCEROL, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Bosanac, I, Yamazaki, H, Matsu-ura, T, Michikawa, T, Mikoshiba, K, Ikura, M. | | Deposit date: | 2004-11-13 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the ligand binding suppressor domain of type 1 inositol 1,4,5-trisphosphate receptor.
Mol.Cell, 17, 2005
|
|
1Y1A
 
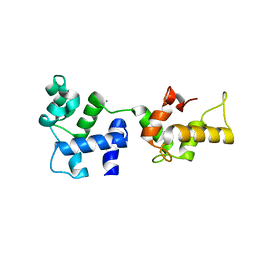 | | CRYSTAL STRUCTURE OF CALCIUM AND INTEGRIN BINDING PROTEIN | | Descriptor: | CALCIUM ION, Calcium and integrin binding 1 (calmyrin), GLUTATHIONE | | Authors: | Blamey, C.J, Ceccarelli, C, Naik, U.P, Bahnson, B.J. | | Deposit date: | 2004-11-17 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of calcium- and integrin-binding protein 1: Insights into redox regulated functions
Protein Sci., 14, 2005
|
|
2B1O
 
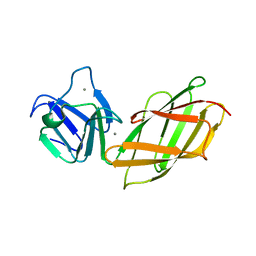 | | Solution Structure of Ca2+-bound DdCAD-1 | | Descriptor: | CALCIUM ION, Calcium-dependent cell adhesion molecule-1 | | Authors: | Lin, Z, Sriskanthadevan, S, Huang, H.B, Siu, C.H, Yang, D.W. | | Deposit date: | 2005-09-16 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the adhesion molecule DdCAD-1 reveal new insights into Ca(2+)-dependent cell-cell adhesion
Nat.Struct.Mol.Biol., 13, 2006
|
|
2AQX
 
 | | Crystal Structure of the Catalytic and CaM-Binding domains of Inositol 1,4,5-Trisphosphate 3-Kinase B | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PREDICTED: inositol 1,4,5-trisphosphate 3-kinase B | | Authors: | Chamberlain, P.P, Sandberg, M.L, Sauer, K, Cooke, M.P, Lesley, S.A, Spraggon, G. | | Deposit date: | 2005-08-18 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into enzyme regulation for inositol 1,4,5-trisphosphate 3-kinase B
Biochemistry, 44, 2005
|
|
2B6P
 
 | | X-ray structure of lens Aquaporin-0 (AQP0) (lens MIP) in an open pore state | | Descriptor: | Lens fiber major intrinsic protein | | Authors: | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | Deposit date: | 2005-10-03 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
2B6O
 
 | | Electron crystallographic structure of lens Aquaporin-0 (AQP0) (lens MIP) at 1.9A resolution, in a closed pore state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Lens fiber major intrinsic protein | | Authors: | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | Deposit date: | 2005-10-03 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.9 Å) | | Cite: | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
2SCP
 
 | |
2SAS
 
 | |
2XOA
 
 | |
2IF4
 
 | |
2KBI
 
 | |
