3ENI
 
 | | Crystal structure of the Fenna-Matthews-Olson Protein from Chlorobaculum Tepidum | | Descriptor: | BACTERIOCHLOROPHYLL A, Bacteriochlorophyll a protein | | Authors: | Tronrud, D, Camara-Artigas, A, Blankenship, R, Allen, J.P. | | Deposit date: | 2008-09-25 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria.
Photosynth.Res., 100, 2009
|
|
6PL3
 
 | | TRK-A IN COMPLEX WITH LIGAND 2a | | Descriptor: | 2-[(3-tert-butyl-1-phenyl-1H-pyrazol-5-yl)amino]-2-oxoethyl 4-(1H-tetrazol-1-yl)benzoate, High affinity nerve growth factor receptor | | Authors: | Subramanian, G. | | Deposit date: | 2019-06-30 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Type 2 inhibitor leads of human tropomyosin receptor kinase (hTrkA).
Bioorg.Med.Chem.Lett., 29, 2019
|
|
4Y1Q
 
 | | Crystal Structure of HasA mutant Y75A monomer from Yersinia pseudotuberculosis | | Descriptor: | Extracellular heme acquisition hemophore HasA, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Hino, T, Kanadani, M, Ozaki, S, Sato, T. | | Deposit date: | 2015-02-08 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of heme acquisition system A from Yersinia pseudotuberculosis (HasAypt): Roles of the axial ligand Tyr75 and two distal arginines in heme binding
J.Inorg.Biochem., 151, 2015
|
|
1K0I
 
 | | Pseudomonas aeruginosa phbh R220Q in complex with 100mM PHB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID, ... | | Authors: | Wang, J, Ortiz-Maldonado, M, Entsch, B, Ballou, D, Gatti, D.L. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein and ligand dynamics in 4-hydroxybenzoate hydroxylase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5C9J
 
 | | Human CD1c with ligands in A' and F' channel | | Descriptor: | Beta-2-microglobulin, GLYCEROL, LAURIC ACID, ... | | Authors: | Tews, I, Machelett, M.M, Mansour, S, Gadola, S.D. | | Deposit date: | 2015-06-27 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cholesteryl esters stabilize human CD1c conformations for recognition by self-reactive T cells.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4N90
 
 | |
8JWV
 
 | | Untethered R0RBR | | Descriptor: | BARIUM ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Lenka, D.R, Kumar, A. | | Deposit date: | 2023-06-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Additional feedforward mechanism of Parkin activation via binding of phospho-UBL and RING0 in trans.
Elife, 13, 2024
|
|
4JSA
 
 | | Benzenesulfonamide complexed with hCAII H94D | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, SULFATE ION, ... | | Authors: | Martin, D.P, Hann, Z.S, Cohen, S.M. | | Deposit date: | 2013-03-22 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Metalloprotein-Inhibitor Binding: Human Carbonic Anhydrase II as a Model for Probing Metal-Ligand Interactions in a Metalloprotein Active Site.
Inorg.Chem., 52, 2013
|
|
5T5U
 
 | |
4Y4S
 
 | | Crystal Structure of Y75A HasA dimer from Yersinia pseudotuberculosis | | Descriptor: | Extracellular heme acquisition hemophore HasA, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Hino, T, Kanadani, M, Muroki, T, Ishimaru, Y, Wada, Y, Sato, T, Ozaki, S. | | Deposit date: | 2015-02-11 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of heme acquisition system A from Yersinia pseudotuberculosis (HasAypt): Roles of the axial ligand Tyr75 and two distal arginines in heme binding
J.Inorg.Biochem., 151, 2015
|
|
1HSR
 
 | | BINDING MODE OF BENZHYDROXAMIC ACID TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BENZHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1997-07-01 | | Release date: | 1998-07-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding mode of benzhydroxamic acid to Arthromyces ramosus peroxidase shown by X-ray crystallographic analysis of the complex at 1.6 A resolution.
FEBS Lett., 412, 1997
|
|
4RZF
 
 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, S441W mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Tian, X, Li, A, Li, L, Liu, Y, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
6MS7
 
 | | Peroxisome proliferator-activated receptor gamma ligand binding domain in complex with a novel selective PPAR-gamma modulator VSP-77 | | Descriptor: | PGC1 LXXLL motif, Peroxisome proliferator-activated receptor gamma, {[(1S)-1-(4-chlorophenyl)octyl]oxy}acetic acid | | Authors: | Yi, W, Jiang, H, Zhou, X.E, Shi, J, Zhao, G, Zhang, X, Sun, Y, Suino-Powell, K, Li, J, Li, J, Melcher, K, Xu, H.E. | | Deposit date: | 2018-10-16 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Identification and structural insight of an effective PPAR gamma modulator with improved therapeutic index for anti-diabetic drug discovery.
Chem Sci, 11, 2020
|
|
4RZE
 
 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, L437W,D594E mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Fengwei, L, Xuyang, T, Anzhong, L, Li, L, Yuan, L, Hangzi, C, Qiao, W, Tianwei, L. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
3TCF
 
 | | Crystal structure of E. coli OppA complexed with endogenous ligands | | Descriptor: | Endogenous peptide, Periplasmic oligopeptide-binding protein | | Authors: | Klepsch, M.M, Kovermann, M, Low, C, Balbach, J, de Gier, J.W, Slotboom, D.J, Berntsson, R.P.-A. | | Deposit date: | 2011-08-09 | | Release date: | 2011-10-12 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Escherichia coli peptide binding protein OppA has a preference for positively charged peptides.
J.Mol.Biol., 414, 2011
|
|
1RYW
 
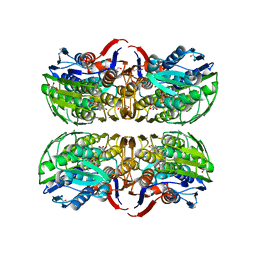 | | C115S MurA liganded with reaction products | | Descriptor: | GLYCEROL, PHOSPHATE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Eschenburg, S, Schonbrunn, E. | | Deposit date: | 2003-12-22 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evidence That the Fosfomycin Target Cys115 in UDP-N-acetylglucosamine Enolpyruvyl Transferase (MurA) Is Essential for Product Release.
J.Biol.Chem., 280, 2005
|
|
4ZFZ
 
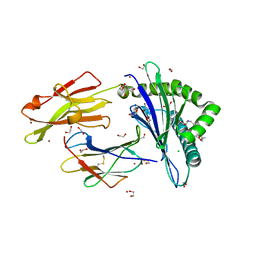 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*098 complexed with myristoylated 5-mer lipopeptide derived from SIV Nef protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-mer lipopeptide from Protein Nef, ... | | Authors: | Morita, D, Sugita, M. | | Deposit date: | 2015-04-22 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Crystal structure of the N-myristoylated lipopeptide-bound MHC class I complex
Nat Commun, 7, 2016
|
|
6QGA
 
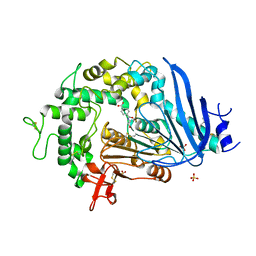 | | Crystal structure of Ideonella sakaiensis MHETase bound to the non-hydrolyzable ligand MHETA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-hydroxyethylcarbamoyl)benzoic acid, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
4JSW
 
 | | Human carbonic anhydrase II H94C | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Martin, D.P, Hann, Z.S, Cohen, S.M. | | Deposit date: | 2013-03-22 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Metalloprotein-Inhibitor Binding: Human Carbonic Anhydrase II as a Model for Probing Metal-Ligand Interactions in a Metalloprotein Active Site.
Inorg.Chem., 52, 2013
|
|
6QGC
 
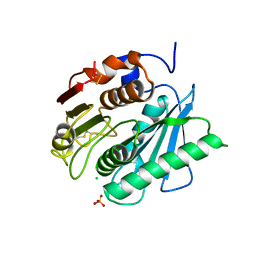 | | PETase from Ideonella sakaiensis without ligand | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
4JSS
 
 | | Human carbonic anhydrase II H94D bound to a bidentate inhibitor | | Descriptor: | 1-hydroxy-2-sulfanylpyridinium, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Martin, D.P, Hann, Z.S, Cohen, S.M. | | Deposit date: | 2013-03-22 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Metalloprotein-Inhibitor Binding: Human Carbonic Anhydrase II as a Model for Probing Metal-Ligand Interactions in a Metalloprotein Active Site.
Inorg.Chem., 52, 2013
|
|
6QI4
 
 | | NCS-1 bound to a ligand | | Descriptor: | 2-(1~{H}-indol-3-yl)-~{N}-[(~{E})-(4-nitro-3-oxidanyl-phenyl)methylideneamino]ethanamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Sanchez-Barrena, M.J, Blanco-Gabella, P. | | Deposit date: | 2019-01-17 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Insights into real-time chemical processes in a calcium sensor protein-directed dynamic library.
Nat Commun, 10, 2019
|
|
3N0Y
 
 | | Adenylate cyclase class IV with active site ligand APC | | Descriptor: | Adenylate cyclase 2, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MANGANESE (II) ION | | Authors: | Gallagher, D.T, Reddy, P.T. | | Deposit date: | 2010-05-14 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Active-Site Structure of Class IV Adenylyl Cyclase and Transphyletic Mechanism.
J.Mol.Biol., 405, 2011
|
|
4RZG
 
 | | Crystal Structure Analysis of the DNPA-bounded NUR77 Ligand binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, pentyl (3,5-dihydroxy-2-nonanoylphenyl)acetate | | Authors: | Li, F, Tian, X, Li, A, Li, L, Liu, Y, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
1C5B
 
 | | DECARBOXYLASE CATALYTIC ANTIBODY 21D8 UNLIGANDED FORM | | Descriptor: | CHIMERIC DECARBOXYLASE ANTIBODY 21D8 | | Authors: | Hotta, K, Wilson, I.A. | | Deposit date: | 1999-11-08 | | Release date: | 2000-10-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalysis of decarboxylation by a preorganized heterogeneous microenvironment: crystal structures of abzyme 21D8.
J.Mol.Biol., 302, 2000
|
|
