4CXQ
 
 | | Mycobaterium tuberculosis transaminase BioA complexed with substrate KAPA | | Descriptor: | 1,2-ETHANEDIOL, 7-KETO-8-AMINOPELARGONIC ACID, ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, ... | | Authors: | Dai, R, Wilson, D.J, Geders, T.W, Aldrich, C.C, Finzel, B.C. | | Deposit date: | 2014-04-08 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of Mycobacterium Tuberculosis Transaminase Bioa by Aryl Hydrazines and Hydrazides.
Chembiochem, 15, 2014
|
|
6VGU
 
 | |
8FHT
 
 | | Cryo-EM structure of human NCC | | Descriptor: | CHLORIDE ION, SODIUM ION, Solute carrier family 12 member 3 | | Authors: | Zhang, J, Fan, M, Feng, L. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structure and thiazide inhibition mechanism of the human Na-Cl cotransporter.
Nature, 614, 2023
|
|
5TMB
 
 | | Crystal structure of Os79 from O. sativa in complex with UDP. | | Descriptor: | Glycosyltransferase, Os79, URIDINE-5'-DIPHOSPHATE | | Authors: | Wetterhorn, K.M, Newmister, S.A, Caniza, R.K, Busman, M, McCormick, S.P, Berthiller, F, Adam, G, Rayment, I. | | Deposit date: | 2016-10-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of Os79 (Os04g0206600) from Oryza sativa: A UDP-glucosyltransferase Involved in the Detoxification of Deoxynivalenol.
Biochemistry, 55, 2016
|
|
4D03
 
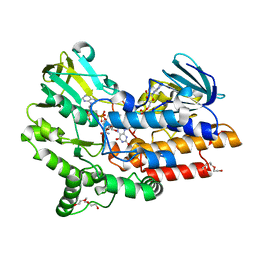 | | Structure of the Cys65Asp mutant of phenylacetone monooxygenase: oxidised state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HEXAETHYLENE GLYCOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Brondani, P.B, Dudek, H.M, Martinoli, C, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2014-04-24 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Finding the Switch: Turning a Baeyer-Villiger Monooxygenase Into a Nadph Oxidase.
J.Am.Chem.Soc., 136, 2014
|
|
4CPI
 
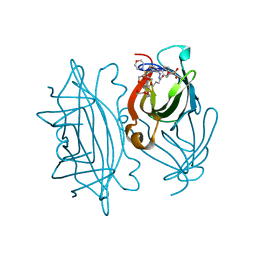 | | streptavidin A86D mutant with love-hate ligand 4 | | Descriptor: | 5-[(3aS,4S,6aR)-2-oxo-hexahydro-1H-thieno[3,4- d]imidazolidin-4-yl]-N'-{2,6-bis[4-(morpholine-4- sulfonyl)phenyl]phenyl}pentanehydrazide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
8FHQ
 
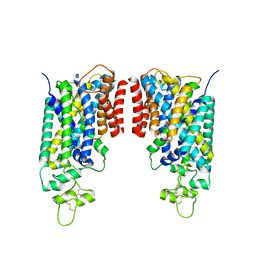 | | Cryo-EM structure of human NCC (class 3-2) | | Descriptor: | Polythiazide, SODIUM ION, Solute carrier family 12 member 2,Solute carrier family 12 member 3 chimera | | Authors: | Zhang, J, Fan, M, Feng, L. | | Deposit date: | 2022-12-14 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structure and thiazide inhibition mechanism of the human Na-Cl cotransporter.
Nature, 614, 2023
|
|
2JOC
 
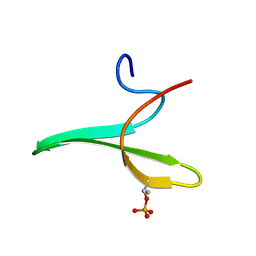 | | Mouse Itch 3rd domain phosphorylated in T30 | | Descriptor: | Itchy E3 ubiquitin protein ligase | | Authors: | Macias, M.J, Shaw, A.Z, Martin-Malpartida, P, Morales, B, Ruiz, L, Ramirez-Espain, X, Yraola, F, Royo, M. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structural Studies of the ItchWW3 Domain Reveal that Phosphorylation at T30 Inhibits the Interaction with PPxY-Containing Ligands
Structure, 15, 2007
|
|
8FRC
 
 | |
6UVC
 
 | | Crystal structure of BCL-XL bound to compound 8: (R)-3-(Benzylthio)-2-(3-(2-((4'-chloro-[1,1'-biphenyl]-2-yl)methyl)-1,2,3,4-tetrahydroisoquinoline-6-carbonyl)-3-(4-methylbenzyl)ureido)propanoic acid | | Descriptor: | (R)-3-(Benzylthio)-2-(3-(2-((4'-chloro-[1,1'-biphenyl]-2-yl)methyl)-1,2,3,4-tetrahydroisoquinoline-6-carbonyl)-3-(4-methylbenzyl)ureido)propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1, ... | | Authors: | Roy, M.J, Birkinshaw, R, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
6UVE
 
 | | Crystal structure of BCL-XL bound to compound 7: (R)-3-(Benzylthio)-2-(3-(4-chloro-[1,1':2',1'':3'',1'''-quaterphenyl]-4'''-carbonyl)-3-(4-methylbenzyl)ureido)propanoic acid | | Descriptor: | (R)-3-(Benzylthio)-2-(3-(4-chloro-[1,1':2',1'':3'',1'''-quaterphenyl]-4'''-carbonyl)-3-(4-methylbenzyl)ureido)propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1 | | Authors: | Roy, M.J, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
6UVD
 
 | | Crystal structure of BCL-XL bound to compound 2: (2R)-3-(Benzylsulfanyl)-2-({[(4-methylphenyl)methyl] [(4 phenylphenyl)carbonyl] carbamoyl}amino) propanoic acid | | Descriptor: | (2R)-3-(Benzylsulfanyl)-2-({[(4-methylphenyl)methyl] [(4 phenylphenyl)carbonyl] carbamoyl}amino) propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1, ... | | Authors: | Roy, M.J, Birkinshaw, R, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
6UVH
 
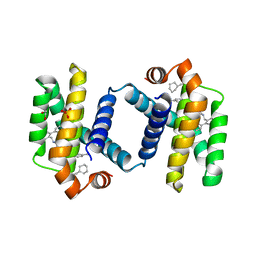 | | Crystal structure of BCL-XL bound to compound 15: (R)-2-(3-(2-((4'-Chloro-[1,1'-biphenyl]-2-yl)methyl)-1,2,3,4-tetrahydroisoquinoline-6-carbonyl)-3-(4-methylbenzyl)ureido)-3-((cyclohexylmethyl)sulfonyl)propanoic acid | | Descriptor: | (R)-2-(3-(2-((4'-Chloro-[1,1'-biphenyl]-2-yl)methyl)-1,2,3,4-tetrahydroisoquinoline-6-carbonyl)-3-(4-methylbenzyl)ureido)-3-((cyclohexylmethyl)sulfonyl)propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1, ... | | Authors: | Roy, M.J, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
8FRA
 
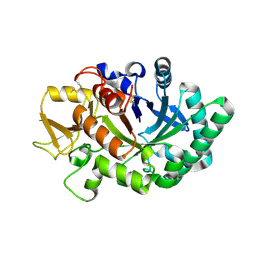 | | Mouse acidic mammalian chitinase, catalytic domain in complex with diacetylchitobiose at pH 5.60 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acidic mammalian chitinase, ... | | Authors: | Diaz, R.E, Fraser, J.S. | | Deposit date: | 2023-01-06 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase.
Biorxiv, 2024
|
|
6UVF
 
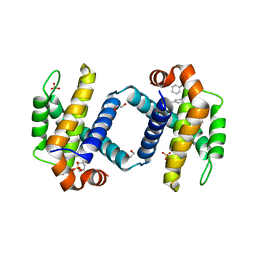 | | Crystal structure of BCL-XL bound to compound 12: (R)-2-(3-([1,1'-Biphenyl]-4-carbonyl)-3-(4-methylbenzyl)ureido)-3-((cyclohexylmethyl)sulfonyl)propanoic acid | | Descriptor: | (R)-2-(3-([1,1'-Biphenyl]-4-carbonyl)-3-(4-methylbenzyl)ureido)-3-((cyclohexylmethyl)sulfonyl)propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1, ... | | Authors: | Roy, M.J, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
6UVG
 
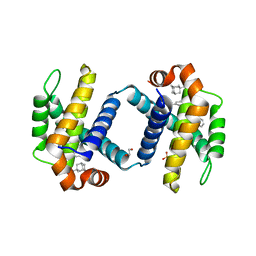 | | Crystal structure of BCL-XL bound to compound 13: (R)-2-(3-([1,1'-Biphenyl]-4-carbonyl)-3-(4-methylbenzyl)ureido)-3-(((3R,5R,7R)-adamantan-1-ylmethyl)sulfonyl)propanoic acid | | Descriptor: | (R)-2-(3-([1,1'-Biphenyl]-4-carbonyl)-3-(4-methylbenzyl)ureido)-3-(((3R,5R,7R)-adamantan-1-ylmethyl)sulfonyl)propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1, ... | | Authors: | Roy, M.J, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
8FYN
 
 | | MicroED structure of A2A from plasma milled lamellae | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Martynowycz, M.W, Shiriaeva, A, Clabbers, M.T.B, Nicolas, W.J, Weaver, S.J, Hattne, J, Gonen, T. | | Deposit date: | 2023-01-26 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | A robust approach for MicroED sample preparation of lipidic cubic phase embedded membrane protein crystals.
Nat Commun, 14, 2023
|
|
5U9F
 
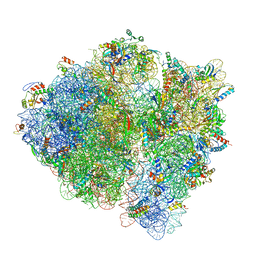 | | 3.2 A cryo-EM ArfA-RF2 ribosome rescue complex (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Svidritskiy, E, Madireddy, R, Diaz-Avalos, R, Grant, T, Grigorieff, N, Sousa, D, Korostelev, A.A. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of ribosome rescue by ArfA and RF2.
Elife, 6, 2017
|
|
5TZY
 
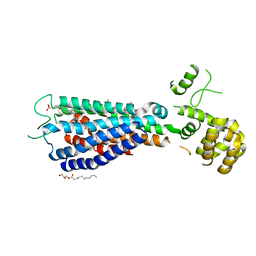 | | GPR40 in complex with AgoPAM AP8 and partial agonist MK-8666 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3R)-3-cyclopropyl-3-[(2R)-2-(1-{(1S)-1-[5-fluoro-2-(trifluoromethoxy)phenyl]ethyl}piperidin-4-yl)-3,4-dihydro-2H-1-benzopyran-7-yl]-2-methylpropanoic acid, (5aR,6S,6aS)-3-({2',6'-dimethyl-4'-[3-(methylsulfonyl)propoxy][1,1'-biphenyl]-3-yl}methoxy)-5,5a,6,6a-tetrahydrocyclopropa[4,5]cyclopenta[1,2-c]pyridine-6-carboxylic acid, ... | | Authors: | Lu, J, Byrne, N, Patel, S, Sharma, S, Soisson, S.M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40.
Nat. Struct. Mol. Biol., 24, 2017
|
|
2JQY
 
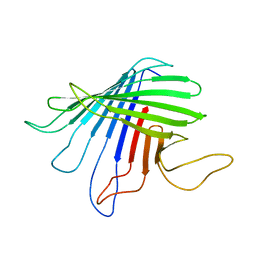 | | Outer Membrane Protein G | | Descriptor: | Outer membrane protein G | | Authors: | Liang, B, Tamm, L.K. | | Deposit date: | 2007-06-15 | | Release date: | 2007-10-09 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of outer membrane protein G by solution NMR spectroscopy.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
4D3V
 
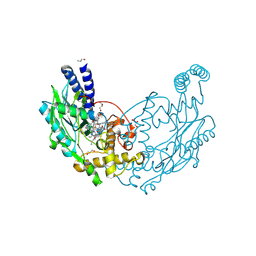 | |
4CIN
 
 | | Complex of Bcl-xL with its BH3 domain | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Colman, P.M, Lee, E.F, Fairlie, W.D. | | Deposit date: | 2013-12-12 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | The Functional Differences of Pro-Survival and Pro-Apoptotic B Cell Lymphoma 2 (Bcl-2) Proteins Depend on Structural Differences in Their Bcl-2 Homology 3 (Bh3) Domains
J.Biol.Chem., 289, 2014
|
|
6REQ
 
 | |
5U9G
 
 | | 3.2 A cryo-EM ArfA-RF2 ribosome rescue complex (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Svidritskiy, E, Madireddy, R, Diaz-Avalos, R, Grant, T, Grigorieff, N, Sousa, D, Korostelev, A.A. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of ribosome rescue by ArfA and RF2.
Elife, 6, 2017
|
|
4D3K
 
 | | Structure of Bacillus subtilis nitric oxide synthase in complex with 6,6'-((5-(3-aminopropyl)-1,3-phenylene)bis(ethane-2,1-diyl))bis(4- methylpyridin-2-amine) | | Descriptor: | 6,6'-{[5-(3-aminopropyl)benzene-1,3-diyl]diethane-2,1-diyl}bis(4-methylpyridin-2-amine), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.017 Å) | | Cite: | Structure-Based Design of Bacterial Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 58, 2015
|
|
