1CUL
 
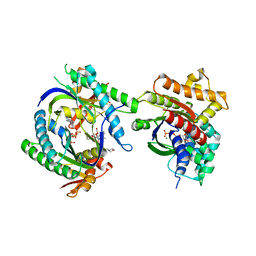 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH 2',5'-DIDEOXY-ADENOSINE 3'-TRIPHOSPHATE AND MG | | Descriptor: | 2',5'-DIDEOXY-ADENOSINE 3'-MONOPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Dessauer, C.A, Sunahara, R.K, Johnson, R.A, Gilman, A.G, Sprang, S.R. | | Deposit date: | 1999-08-20 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for P-site inhibition of adenylyl cyclase.
Biochemistry, 39, 2000
|
|
1CS4
 
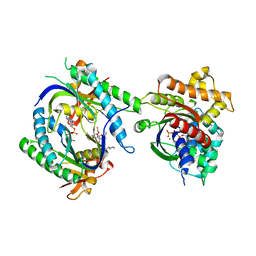 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH 2'-DEOXY-ADENOSINE 3'-MONOPHOSPHATE, PYROPHOSPHATE AND MG | | Descriptor: | 2'-DEOXY-ADENOSINE 3'-MONOPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Dessauer, C.A, Sunahara, R.K, Johnson, R.A, Gilman, A.G, Sprang, S.R. | | Deposit date: | 1999-08-16 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for P-site inhibition of adenylyl cyclase.
Biochemistry, 39, 2000
|
|
1C3U
 
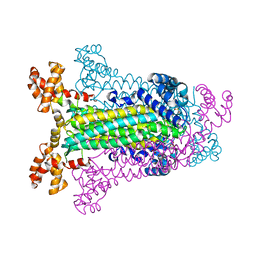 | | T. MARITIMA ADENYLOSUCCINATE LYASE | | Descriptor: | ADENYLOSUCCINATE LYASE, SULFATE ION | | Authors: | Toth, E.A, Yeates, T.O. | | Deposit date: | 1999-07-28 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of adenylosuccinate lyase, an enzyme with dual activity in the de novo purine biosynthetic pathway.
Structure Fold.Des., 8, 2000
|
|
5DJF
 
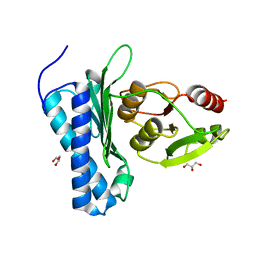 | |
5DJG
 
 | | Structure of M. tuberculosis CysQ, a PAP phosphatase with PAP, Mg, and Li bound | | Descriptor: | 3'-phosphoadenosine 5'-phosphate phosphatase, ADENOSINE-3'-5'-DIPHOSPHATE, LITHIUM ION, ... | | Authors: | Fisher, A.J, Erickson, A.I. | | Deposit date: | 2015-09-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Crystal Structures of Mycobacterium tuberculosis CysQ, with Substrate and Products Bound.
Biochemistry, 54, 2015
|
|
5DJJ
 
 | | Structure of M. tuberculosis CysQ, a PAP phosphatase with PO4 and 2Mg bound | | Descriptor: | 3'-phosphoadenosine 5'-phosphate phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fisher, A.J, Erickson, A.I. | | Deposit date: | 2015-09-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal Structures of Mycobacterium tuberculosis CysQ, with Substrate and Products Bound.
Biochemistry, 54, 2015
|
|
5DJK
 
 | | Structure of M. tuberculosis CysQ, a PAP phosphatase with PO4 and 2Ca bound | | Descriptor: | 3'-phosphoadenosine 5'-phosphate phosphatase, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Fisher, A.J, Erickson, A.I. | | Deposit date: | 2015-09-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal Structures of Mycobacterium tuberculosis CysQ, with Substrate and Products Bound.
Biochemistry, 54, 2015
|
|
7DQ6
 
 | | Crystal structure of HitB in complex with (S)-beta-3-Br-phenylalanine sulfamoyladenosine | | Descriptor: | CALCIUM ION, Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl N-[(3S)-3-azanyl-3-(3-bromophenyl)propanoyl]sulfamate | | Authors: | Kudo, F, Takahashi, S, Miyanaga, A, Nakazawa, Y, Eguchi, T. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutational Biosynthesis of Hitachimycin Analogs Controlled by the beta-Amino Acid-Selective Adenylation Enzyme HitB.
Acs Chem.Biol., 16, 2021
|
|
7DQ5
 
 | | Crystal structure of HitB in complex with (S)-beta-phenylalanine sulfamoyladenosine | | Descriptor: | CALCIUM ION, Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[(3~{S})-3-azanyl-3-phenyl-propanoyl]sulfamate | | Authors: | Kudo, F, Takahashi, S, Miyanaga, A, Nakazawa, Y, Eguchi, T. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mutational Biosynthesis of Hitachimycin Analogs Controlled by the beta-Amino Acid-Selective Adenylation Enzyme HitB.
Acs Chem.Biol., 16, 2021
|
|
7S0Y
 
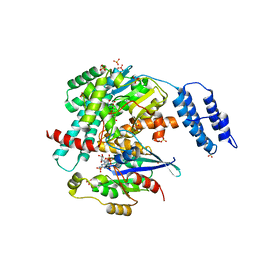 | | Structures of TcdB in complex with Cdc42 | | Descriptor: | Cell division control protein 42 homolog, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zheng, L, Rongsheng, J, Peng, C. | | Deposit date: | 2021-08-31 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis for selective modification of Rho and Ras GTPases by Clostridioides difficile toxin B.
Sci Adv, 7, 2021
|
|
1JDE
 
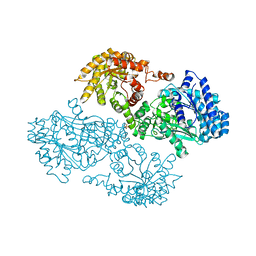 | | K22A mutant of pyruvate, phosphate dikinase | | Descriptor: | PYRUVATE, PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Ye, D, Wei, M, McGuire, M, Huang, K, Kapadia, G, Herzberg, O, Martin, B.M, Dunaway-Mariano, D. | | Deposit date: | 2001-06-13 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Investigation of the catalytic site within the ATP-grasp domain of Clostridium symbiosum pyruvate phosphate dikinase.
J.Biol.Chem., 276, 2001
|
|
5D1O
 
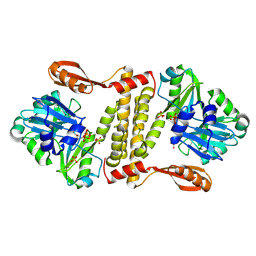 | | Archaeal ATP-dependent RNA ligase - form 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA ligase, MAGNESIUM ION, ... | | Authors: | Murakami, K.S. | | Deposit date: | 2015-08-04 | | Release date: | 2016-03-09 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Structural and mutational analysis of archaeal ATP-dependent RNA ligase identifies amino acids required for RNA binding and catalysis.
Nucleic Acids Res., 44, 2016
|
|
5D1P
 
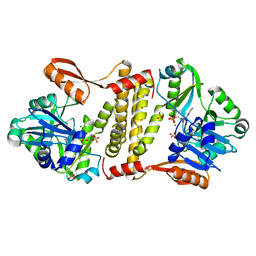 | | Archaeal ATP-dependent RNA ligase - form 2 | | Descriptor: | ATP-dependent RNA ligase, MAGNESIUM ION, SULFATE ION | | Authors: | Murakami, K.S. | | Deposit date: | 2015-08-04 | | Release date: | 2016-03-09 | | Last modified: | 2016-03-30 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structural and mutational analysis of archaeal ATP-dependent RNA ligase identifies amino acids required for RNA binding and catalysis.
Nucleic Acids Res., 44, 2016
|
|
8GI6
 
 | | Crystal structure of RhoA mutant L69R complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Chen, X, Qian, X, Chandravanshi, M, Lowy, D.R, Walters, K.J. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ras-like GTPases mutant structures
To be published
|
|
8GI3
 
 | | Crystal structure of RhoA mutant L69P complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Chen, X, Qian, X, Chandravanshi, M, Lowy, D.R, Walters, K.J. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Ras-like GTPases mutants structure
To be published
|
|
2DIK
 
 | | R337A MUTANT OF PYRUVATE PHOSPHATE DIKINASE | | Descriptor: | PROTEIN (PYRUVATE PHOSPHATE DIKINASE), SULFATE ION | | Authors: | Huang, K, Herzberg, O. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Location of the phosphate binding site within Clostridium symbiosum pyruvate phosphate dikinase.
Biochemistry, 37, 1998
|
|
7CBA
 
 | |
5IFI
 
 | | CRYSTAL STRUCTURE OF ACETYL-COA SYNTHETASE IN COMPLEX WITH ADENOSINE-5'-PROPYLPHOSPHATE FROM CRYPTOCOCCUS NEOFORMANS H99 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-MONOPHOSPHATE-PROPYL ESTER, Acetyl-coenzyme A synthetase, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), SSGCID, Fox III, D, Edwards, T.E, Lorimer, D.D, Mutz, M.W. | | Deposit date: | 2016-02-26 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Acetyl-CoA Synthetase in complex with Adenosine-5'-propylphosphate from Cryptococcus neoformans H99
To Be Published
|
|
5K85
 
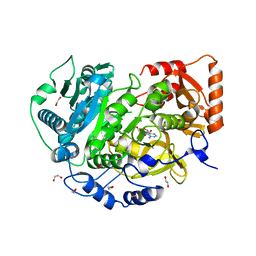 | | Crystal Structure of Acetyl-CoA Synthetase in Complex with Adenosine-5'-propylphosphate and Coenzyme A from Cryptococcus neoformans H99 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-MONOPHOSPHATE-PROPYL ESTER, Acetyl-coenzyme A synthetase, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Fox III, D, Delker, S.L, Potts, K.T, Lorimer, D.D, Edwards, T.E, Mutz, M.W, SSGCID | | Deposit date: | 2016-05-27 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF ACETYL-COA SYNTHETASE IN COMPLEX WITH ADENOSINE-5'-PROPYLPHOSPHATE AND COENZYME A FROM CRYPTOCOCCUS NEOFORMANS H99
To Be Published
|
|
3DZQ
 
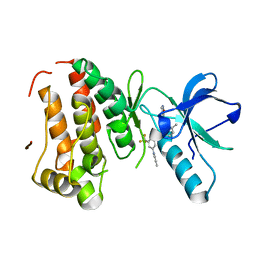 | | Human EphA3 kinase domain in complex with inhibitor AWL-II-38.3 | | Descriptor: | EPH receptor A3, N-[2-methyl-5-({[3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl]carbonyl}amino)phenyl]isoxazole-5-carboxamide | | Authors: | Walker, J.R, Syeda, F, Gray, N, Mansoor, W, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-30 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kinase Domain of Human Ephrin Type-A Receptor 3 (Epha3) in Complex with ALW-II-38-3.
To be Published
|
|
2J6M
 
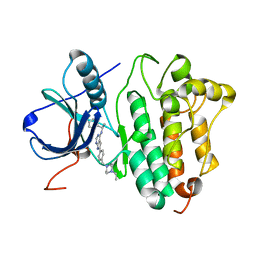 | | Crystal structure of EGFR kinase domain in complex with AEE788 | | Descriptor: | 6-{4-[(4-ETHYLPIPERAZIN-1-YL)METHYL]PHENYL}-N-[(1R)-1-PHENYLETHYL]-7H-PYRROLO[2,3-D]PYRIMIDIN-4-AMINE, EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-09-29 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2J5F
 
 | |
2IY5
 
 | | PHENYLALANYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS complexed with tRNA and a phenylalanyl-adenylate analog | | Descriptor: | ADENOSINE-5'-[PHENYLALANINOL-PHOSPHATE], MAGNESIUM ION, PHENYLALANYL-TRNA SYNTHETASE ALPHA CHAIN, ... | | Authors: | Moor, N, Kotik-Kogan, O, Tworowski, D, Sukhanova, M, Safro, M. | | Deposit date: | 2006-07-12 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The crystal structure of the ternary complex of phenylalanyl-tRNA synthetase with tRNAPhe and a phenylalanyl-adenylate analogue reveals a conformational switch of the CCA end.
Biochemistry, 45, 2006
|
|
2JIU
 
 | | Crystal structure of EGFR kinase domain T790M mutation in complex with AEE788 | | Descriptor: | 6-{4-[(4-ETHYLPIPERAZIN-1-YL)METHYL]PHENYL}-N-[(1R)-1-PHENYLETHYL]-7H-PYRROLO[2,3-D]PYRIMIDIN-4-AMINE, EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Yun, C.-H, Mengwasser, K.E, Toms, A.V, Woo, M.S, Greulich, H, Wong, K.-K, Meyerson, M, Eck, M.J. | | Deposit date: | 2007-07-01 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The T790M Mutation in Egfr Kinase Causes Drug Resistance by Increasing the Affinity for ATP.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2JIT
 
 | | Crystal structure of EGFR kinase domain T790M mutation | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Yun, C.-H, Mengwasser, K.E, Toms, A.V, Woo, M.S, Greulich, H, Wong, K.-K, Meyerson, M, Eck, M.J. | | Deposit date: | 2007-07-01 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The T790M Mutation in Egfr Kinase Causes Drug Resistance by Increasing the Affinity for ATP.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
