9C7J
 
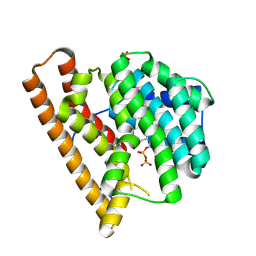 | | Crystal structure of caryolan-1-ol synthase complexed with 2-fluorofarnesyl diphosphate | | Descriptor: | (+)-caryolan-1-ol synthase, (2Z,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, DIPHOSPHATE, ... | | Authors: | Prem Kumar, R, Black, B.Y, Oprian, D.D. | | Deposit date: | 2024-06-10 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of Caryolan-1-ol Synthase, a Sesquiterpene Synthase Catalyzing an Initial Anti-Markovnikov Cyclization Reaction.
Biochemistry, 63, 2024
|
|
7ME8
 
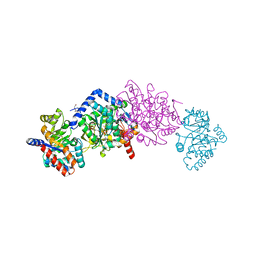 | | The internal aldimine form of the wild-type Salmonella Typhimurium Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor at the beta-site, sodium ion at the metal coordination site and dual beta-Q114 rotamer conformation at 1.60 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The internal aldimine form of the wild-type Salmonella Typhimurium Tryptophan Synthase in complex with N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor at the beta-site, sodium ion at the metal coordination site and dual beta-Q114 rotamer conformation at 1.60 Angstrom resolution.
To be Published
|
|
8K66
 
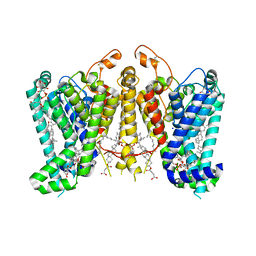 | | Cryo-EM structure of Oryza sativa HKT2;1 at 2.5 angstrom | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Wang, X, Shen, X, Qu, Y, Wang, C, Shen, H. | | Deposit date: | 2023-07-25 | | Release date: | 2024-04-03 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural insights into ion selectivity and transport mechanisms of Oryza sativa HKT2;1 and HKT2;2/1 transporters.
Nat.Plants, 10, 2024
|
|
7UOQ
 
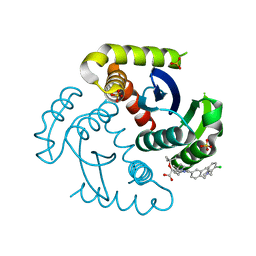 | | CRYSTAL STRUCTURE OF HIV-1 INTEGRASE COMPLEXED WITH (2S)-2-(TERT-BUTOXY)-2-(5-{2-[(2-CHLORO-6-M ETHYLPHENYL)METHYL]-1,2,3,4-TETRAHYDROISOQUINOLIN-6-YL}-4- (4,4-DIMETHYLPIPERIDIN-1-YL)-2-METHYLPYRIDIN-3-YL)ACETIC ACID | | Descriptor: | (2S)-tert-butoxy[(5M)-5-{2-[(2-chloro-6-methylphenyl)methyl]-1,2,3,4-tetrahydroisoquinolin-6-yl}-4-(4,4-dimethylpiperidin-1-yl)-2-methylpyridin-3-yl]acetic acid, Integrase, SULFATE ION | | Authors: | Lewis, H.A, Muckelbauer, J.K. | | Deposit date: | 2022-04-13 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8867 Å) | | Cite: | Discovery and Preclinical Profiling of GSK3839919, a Potent HIV-1 Allosteric Integrase Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
8U6H
 
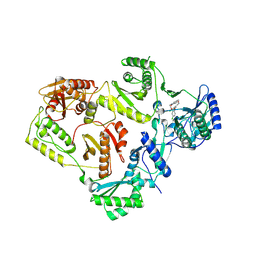 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-(2-(2-(3-acryloyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-1-yl)ethoxy)-4-chlorophenoxy)-5-chlorobenzonitrile (JLJ744), a non-nucleoside inhibitor | | Descriptor: | 3-chloro-5-{4-chloro-2-[2-(2-oxo-3-propanoyl-2,3-dihydro-1H-benzimidazol-1-yl)ethoxy]phenoxy}benzonitrile, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-15 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
5KLZ
 
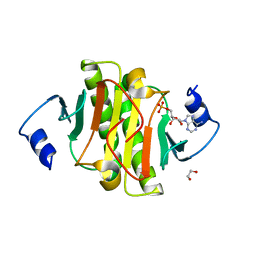 | |
7USY
 
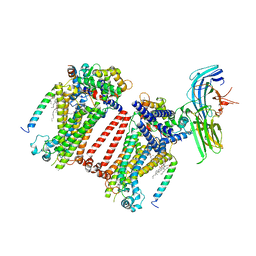 | | Structure of C. elegans TMC-1 complex with ARRD-6 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jeong, H, Clark, S, Gouaux, E. | | Deposit date: | 2022-04-26 | | Release date: | 2022-10-19 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structures of the TMC-1 complex illuminate mechanosensory transduction.
Nature, 610, 2022
|
|
7USX
 
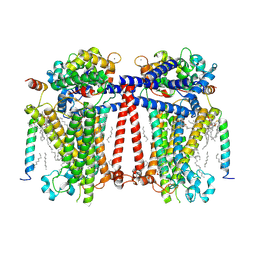 | | Structure of Contracted C. elegans TMC-1 complex | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jeong, H, Clark, S, Gouaux, E. | | Deposit date: | 2022-04-26 | | Release date: | 2022-10-19 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structures of the TMC-1 complex illuminate mechanosensory transduction.
Nature, 610, 2022
|
|
8D0X
 
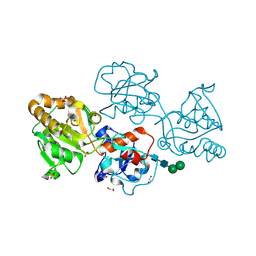 | | Human FUT9 bound to LNnT | | Descriptor: | 1,2-ETHANEDIOL, 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase 9, GLYCEROL, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural basis for Lewis antigen synthesis by the alpha 1,3-fucosyltransferase FUT9.
Nat.Chem.Biol., 19, 2023
|
|
8D0S
 
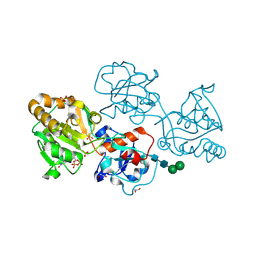 | | Human FUT9 bound to GDP and LNnT | | Descriptor: | 1,2-ETHANEDIOL, 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase 9, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural basis for Lewis antigen synthesis by the alpha 1,3-fucosyltransferase FUT9.
Nat.Chem.Biol., 19, 2023
|
|
8D0R
 
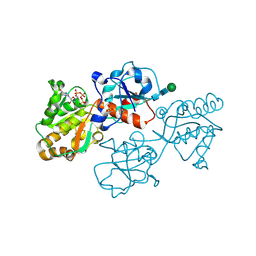 | | Human FUT9 bound to GDP and H-Type 2 | | Descriptor: | 1,2-ETHANEDIOL, 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase 9, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for Lewis antigen synthesis by the alpha 1,3-fucosyltransferase FUT9.
Nat.Chem.Biol., 19, 2023
|
|
7U2U
 
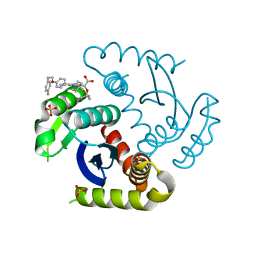 | | CRYSTAL STRUCTURE OF HIV-1 INTEGRASE COMPLEXED WITH Compound-2a AKA (2S)-2-(TERT-BUTOXY)-2-[7-(4,4-DIMETHYLPIPE RIDIN-1-YL)-8-{4-[2-(4-FLUOROPHENYL)ETHOXY]PHENYL}-2,5-DIM ETHYLIMIDAZO[1,2-A]PYRIDIN-6-YL]ACETIC ACID | | Descriptor: | (2S)-tert-butoxy[(4S)-7-(4,4-dimethylpiperidin-1-yl)-8-{4-[2-(4-fluorophenyl)ethoxy]phenyl}-2,5-dimethylimidazo[1,2-a]pyridin-6-yl]acetic acid, Integrase, SULFATE ION | | Authors: | Khan, J.A, lewis, H, Kish, K. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | Scaffold modifications to the 4-(4,4-dimethylpiperidinyl) 2,6-dimethylpyridinyl class of HIV-1 allosteric integrase inhibitors.
Bioorg.Med.Chem., 67, 2022
|
|
7B4W
 
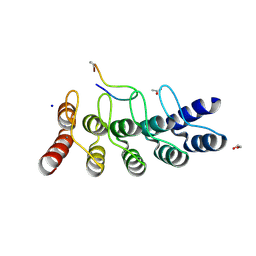 | | Broadly neutralizing DARPin bnD.3 in complex with the HIV-1 envelope variable loop 3 crown mimetic peptide V3-IF (BG505) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Broadly neutralizing DARPin bnD.3, ... | | Authors: | Friedrich, N, Stiegeler, E, Glogl, M, Lemmin, T, Hansen, S, Kadelka, C, Wu, Y, Ernst, P, Maliqi, L, Foulkes, C, Morin, M, Eroglu, M, Liechti, T, Ivan, B, Reinberg, T, Schaefer, J, Karakus, U, Ursprung, S, Mann, A, Rusert, P, Kouyos, R.D, Robinson, J.A, Gunthard, H.F, Pluckthun, A, Trkola, A. | | Deposit date: | 2020-12-02 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
8TD9
 
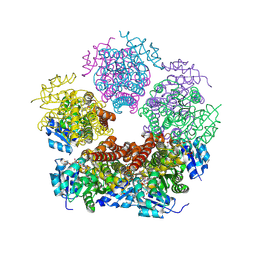 | | Structure of PYCR1 complexed with NADH and L-pipecolic acid | | Descriptor: | (2S)-piperidine-2-carboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
5NJ7
 
 | |
4S0X
 
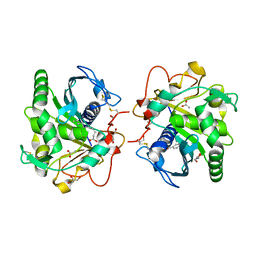 | | Structure of three phase partition - treated lipase from Thermomyces lanuginosa in complex with lauric acid at 2.1 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitrobenzaldehyde, ... | | Authors: | Kumar, M, Mukherjee, J, Gupta, M.N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-01-07 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of three phase partition - treated lipase from Thermomyces lanuginosa in complex with lauric acid at 2.1 A resolution
To be Published
|
|
7NMK
 
 | | Crystal structure of the heterocyclic toxin methyltransferase from Mycobacterium tuberculosis with bound methylation product 1-methoxyquinolin-4(1H)-one | | Descriptor: | 1-methoxy-4-oxoquinoline, 2-heptyl-1-hydroxyquinolin-4(1H)-one methyltransferase, FORMIC ACID, ... | | Authors: | Denkhaus, L, Sartor, P, Einsle, O, Gerhardt, S, Fetzner, S. | | Deposit date: | 2021-02-23 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.204 Å) | | Cite: | Structural basis of O-methylation of (2-heptyl-)1-hydroxyquinolin-4(1H)-one and related compounds by the heterocyclic toxin methyltransferase Rv0560c of Mycobacterium tuberculosis.
J.Struct.Biol., 213, 2021
|
|
6X5T
 
 | | Human Alpha-1,6-fucosyltransferase (FUT8) bound to GDP and A3-Asn | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-(1,6)-fucosyltransferase, GLYCEROL, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Characterizing human alpha-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases.
J.Biol.Chem., 295, 2020
|
|
8ASA
 
 | | Crystal structure of AO75L | | Descriptor: | 1,2-ETHANEDIOL, BICINE, CALCIUM ION, ... | | Authors: | Laugeri, M.E, Speciale, I, Gimeno, A, Lin, S, Poveda, A, Lowary, T, Van Etten, J.L, Barbero, J.J, De Castro, C, Tonetti, M, Rojas, A.L. | | Deposit date: | 2022-08-18 | | Release date: | 2023-08-30 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unveiling the GT114 family: Structural characterization of A075L, a glycosyltransferase from Paramecium bursaria chlorella virus-1 (PBCV-1).
Protein Sci., 33, 2024
|
|
6PBN
 
 | | Pseudopaline Dehydrogenase with (R)-Pseudopaline Soaked 1 hour | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, N-[(3S)-3-amino-3-carboxypropyl]-L-histidine, ... | | Authors: | McFarlane, J.S, Lamb, A.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Staphylopine and pseudopaline dehydrogenase from bacterial pathogens catalyze reversible reactions and produce stereospecific metallophores.
J.Biol.Chem., 294, 2019
|
|
6XP9
 
 | |
8AVQ
 
 | | AO75L in Complex with UDP-Xylose | | Descriptor: | 1,2-ETHANEDIOL, AO75L, BICINE, ... | | Authors: | Laugeri, M.E, Speciale, I, Gimeno, A, Lin, S, Poveda, A, Lowary, T, Van Etten, J.L, Barbero, J.J, De Castro, C, Tonetti, M, Rojas, A.L. | | Deposit date: | 2022-08-26 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unveiling the GT114 family: Structural characterization of A075L, a glycosyltransferase from Paramecium bursaria chlorella virus-1 (PBCV-1).
Protein Sci., 33, 2024
|
|
6SVM
 
 | | Crystal structure of human GFAT-1 in complex with Glucose-6-Phosphate, L-Glu, and UDP-GalNAc | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1, ... | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
8VU9
 
 | | Crystal structure of wild-type HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 5i3 | | Descriptor: | (2E)-3-[4-({2-[(1-{[4-(methanesulfonyl)phenyl]methyl}piperidin-4-yl)amino]pyrido[2,3-d]pyrimidin-4-yl}oxy)-3,5-dimethylphenyl]prop-2-enenitrile, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2024-01-29 | | Release date: | 2025-05-07 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Development of enhanced HIV-1 non-nucleoside reverse transcriptase inhibitors with improved resistance and pharmacokinetic profiles.
Sci Adv, 11, 2025
|
|
7M1R
 
 | |
