3PKJ
 
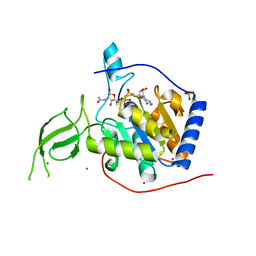 | | Human SIRT6 crystal structure in complex with 2'-N-Acetyl ADP ribose | | 分子名称: | NAD-dependent deacetylase sirtuin-6, SULFATE ION, UNKNOWN ATOM OR ION, ... | | 著者 | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Walker, J.R, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | 登録日 | 2010-11-11 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
5GHY
 
 | |
3PKI
 
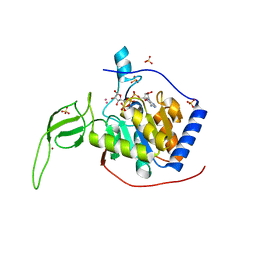 | | Human SIRT6 crystal structure in complex with ADP ribose | | 分子名称: | NAD-dependent deacetylase sirtuin-6, SULFATE ION, UNKNOWN ATOM OR ION, ... | | 著者 | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | 登録日 | 2010-11-11 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
5GHZ
 
 | |
5GHX
 
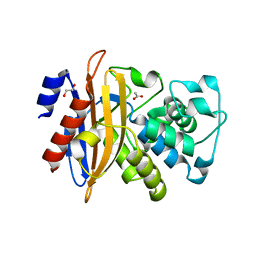 | | Crystal structure of beta-lactamase PenP mutant-E166H | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, Beta-lactamase | | 著者 | Pan, X, Zhao, Y. | | 登録日 | 2016-06-21 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Crystallographic Snapshots of Class A beta-Lactamase Catalysis Reveal Structural Changes That Facilitate beta-Lactam Hydrolysis
J. Biol. Chem., 292, 2017
|
|
5JBV
 
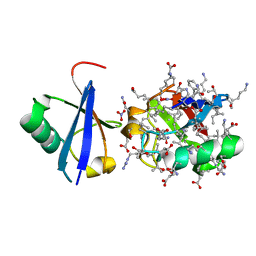 | |
5J8P
 
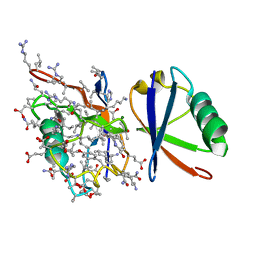 | |
7LN4
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (FOM, Class 3) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Pan, M, Yu, Y, Liu, L, Zhao, M. | | 登録日 | 2021-02-06 | | 公開日 | 2021-09-15 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LMY
 
 | | Cryo-EM structure of human p97 in complex with NMS-873 in the presence of ATP, Npl4/Ufd1, and Ub6 | | 分子名称: | 3-[3-cyclopentylsulfanyl-5-[[3-methyl-4-(4-methylsulfonylphenyl)phenoxy]methyl]-1,2,4-triazol-4-yl]pyridine, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Pan, M, Yu, Y, Liu, L, Zhao, M. | | 登録日 | 2021-02-06 | | 公開日 | 2021-09-15 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN5
 
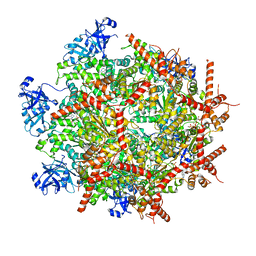 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (CHAPSO, Class 1, Close State) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Pan, M, Yu, Y, Liu, L, Zhao, M. | | 登録日 | 2021-02-06 | | 公開日 | 2021-09-15 | | 実験手法 | ELECTRON MICROSCOPY (3.09 Å) | | 主引用文献 | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN3
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (FOM, Class 2) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Pan, M, Yu, Y, Liu, L, Zhao, M. | | 登録日 | 2021-02-06 | | 公開日 | 2021-09-15 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN0
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and Ub6 (Class 2) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Hexa-ubiquitin, ... | | 著者 | Pan, M, Yu, Y, Liu, L, Zhao, M. | | 登録日 | 2021-02-06 | | 公開日 | 2021-09-15 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN1
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and Ub6 (Class 3) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Hexa-ubiquitin, ... | | 著者 | Pan, M, Yu, Y, Liu, L, Zhao, M. | | 登録日 | 2021-02-06 | | 公開日 | 2021-09-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN2
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (FOM, Class 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Pan, M, Yu, Y, Liu, L, Zhao, M. | | 登録日 | 2021-02-06 | | 公開日 | 2021-09-15 | | 実験手法 | ELECTRON MICROSCOPY (3.63 Å) | | 主引用文献 | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LMZ
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and Ub6 (Class 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Hexa-ubiquitin, ... | | 著者 | Pan, M, Yu, Y, Liu, L, Zhao, M. | | 登録日 | 2021-02-06 | | 公開日 | 2021-09-15 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LN6
 
 | | Cryo-EM structure of human p97 in complex with Npl4/Ufd1 and polyubiquitinated Ub-Eos (CHAPSO, Class 2, Open State) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Pan, M, Yu, Y, Liu, L, Zhao, M. | | 登録日 | 2021-02-06 | | 公開日 | 2021-09-15 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Mechanistic insight into substrate processing and allosteric inhibition of human p97.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MEX
 
 | | Structure of yeast Ubr1 in complex with Ubc2 and N-degron | | 分子名称: | E3 ubiquitin-protein ligase UBR1, N-degron, Ubiquitin, ... | | 著者 | Pan, M, Zheng, Q, Wang, T, Liang, L, Yu, Y, Liu, L, Zhao, M. | | 登録日 | 2021-04-08 | | 公開日 | 2021-11-24 | | 最終更新日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Structural insights into Ubr1-mediated N-degron polyubiquitination.
Nature, 600, 2021
|
|
7MEY
 
 | | Structure of yeast Ubr1 in complex with Ubc2 and monoubiquitinated N-degron | | 分子名称: | 2-(ethylamino)ethane-1-thiol, E3 ubiquitin-protein ligase UBR1, Monoubiquitinated N-degron, ... | | 著者 | Pan, M, Zheng, Q, Wang, T, Liang, L, Yu, Y, Liu, L, Zhao, M. | | 登録日 | 2021-04-08 | | 公開日 | 2021-11-24 | | 最終更新日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (3.67 Å) | | 主引用文献 | Structural insights into Ubr1-mediated N-degron polyubiquitination.
Nature, 600, 2021
|
|
6KHJ
 
 | | Supercomplex for electron transfer | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Pan, X, Cao, D, Xie, F, Zhang, X, Li, M. | | 登録日 | 2019-07-15 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase.
Nat Commun, 11, 2020
|
|
4IFJ
 
 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | 分子名称: | Kelch-like ECH-associated protein 1 | | 著者 | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | 登録日 | 2012-12-14 | | 公開日 | 2014-08-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
To be Published
|
|
4IFN
 
 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | 分子名称: | (1R,2R)-2-{[(1S)-1-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid, kelch-like ECH-associated protein 1 | | 著者 | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | 登録日 | 2012-12-14 | | 公開日 | 2013-12-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
Acta Crystallogr.,Sect.D
|
|
4IFL
 
 | | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes | | 分子名称: | Nrf2 peptide, kelch-like ECH-associated protein 1 | | 著者 | Pan, H, Lin, M, Yang, Y, Callaway, K, Baker, J, Diep, L, Yan, J, Tanaka, K, Zhu, Y.L, Konradi, A.W, Jobling, M, Tam, D, Ren, Z, Cheung, H, Bova, M, Riley, B.E, Yao, N, Artis, D.R. | | 登録日 | 2012-12-14 | | 公開日 | 2013-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of apo Keap1, Keap1-peptide, and Keap1-compound complexes
Acta Crystallogr.,Sect.D
|
|
4JHR
 
 | | An auto-inhibited conformation of LGN reveals a distinct interaction mode between GoLoco motifs and TPR motifs | | 分子名称: | G-protein-signaling modulator 2 | | 著者 | Pan, Z, Zhu, J, Shang, Y, Wei, Z, Jia, M, Xia, C, Wen, W, Wang, W, Zhang, M. | | 登録日 | 2013-03-05 | | 公開日 | 2013-06-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | An autoinhibited conformation of LGN reveals a distinct interaction mode between GoLoco motifs and TPR motifs
Structure, 21, 2013
|
|
8IJD
 
 | | Cryo-EM structure of human HCAR2-Gi complex with MK-6892 | | 分子名称: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Pan, X, Fang, Y. | | 登録日 | 2023-02-27 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8IJA
 
 | | Cryo-EM structure of human HCAR2-Gi complex with niacin | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Pan, X, Fang, Y. | | 登録日 | 2023-02-26 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (2.69 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
