1HJ3
 
 | | Cytochrome cd1 Nitrite Reductase, dioxygen complex | | Descriptor: | GLYCEROL, HEME C, HEME D, ... | | Authors: | Sjogren, T, Hajdu, J. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the bound dioxygen species in the cytochrome oxidase reaction of cytochrome cd1 nitrite reductase.
J. Biol. Chem., 276, 2001
|
|
1HJ5
 
 | | Cytochrome cd1 Nitrite Reductase, reoxidised enzyme | | Descriptor: | GLYCEROL, HEME C, HEME D, ... | | Authors: | Sjogren, T, Hajdu, J. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of the bound dioxygen species in the cytochrome oxidase reaction of cytochrome cd1 nitrite reductase.
J. Biol. Chem., 276, 2001
|
|
7YVB
 
 | | Aplysia californica FaNaC in ligand bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FMRFamide-gated Na+ channel, ... | | Authors: | Chen, Q.F, Liu, F.L, Dang, Y, Feng, H, Zhang, Z, Ye, S. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and mechanism of a neuropeptide-activated channel in the ENaC/DEG superfamily.
Nat.Chem.Biol., 19, 2023
|
|
8P9X
 
 | | Vitamin D receptor complex with Xe4Me agonist ligand | | Descriptor: | (1~{R},3~{S},5~{Z})-5-[(2~{E})-2-[(1~{S},3~{a}~{S},7~{a}~{S})-1,7~{a}-dimethyl-1-(5-methyl-5-oxidanyl-hexa-1,3-diynyl)-2,3,3~{a},5,6,7-hexahydroinden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, ACETATE ION, Nuclear receptor coactivator 2, ... | | Authors: | Rochel, N. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A vitamin D-based strategy overcomes chemoresistance in prostate cancer.
Br.J.Pharmacol., 2024
|
|
6VBG
 
 | | Lactose permease complex with thiodigalactoside and nanobody 9043 | | Descriptor: | Galactoside permease, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose, nanobody 9043, ... | | Authors: | Kumar, H, Stroud, R.M, Kaback, H.R, Finer-Moore, J, Smirnova, I, Kasho, V, Pardon, E, Steyart, J. | | Deposit date: | 2019-12-18 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Diversity in kinetics correlated with structure in nano body-stabilized LacY.
Plos One, 15, 2020
|
|
3BUW
 
 | | Crystal structure of c-Cbl-TKB domain complexed with its binding motif in Syk | | Descriptor: | 13-meric peptide from Tyrosine-protein kinase SYK, E3 ubiquitin-protein ligase CBL | | Authors: | Ng, C, Jackson, R.A, Buschdorf, J.P, Sun, Q, Guy, G.R, Sivaraman, J. | | Deposit date: | 2008-01-03 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates
Embo J., 27, 2008
|
|
4WRF
 
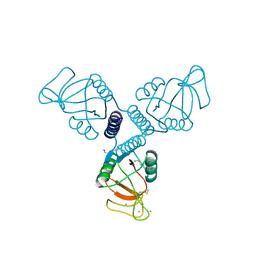 | | Crystal Structure of Surfactant Protein-A DEDN Mutant (E171D/P175E/R197N/K203D) Complexed with Mannose | | Descriptor: | CALCIUM ION, CHLORIDE ION, HEXANE-1,6-DIOL, ... | | Authors: | Rynkiewicz, M.J, Wu, H, Cafarella, T.R, Nikolaidis, N.M, Head, J.F, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2014-10-23 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop
To be published
|
|
6BKE
 
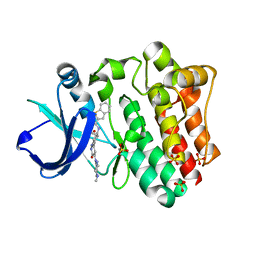 | | BTK complex with compound 10 | | Descriptor: | N-[2-(2-hydroxyethyl)-3-{5-[(5-methyl-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-2-yl)amino]-6-oxo-1,6-dihydropyridazin-3-yl}phenyl]-1-benzothiophene-2-carboxamide, SULFATE ION, Tyrosine-protein kinase BTK | | Authors: | Kiefer, J.R, Eigenbrot, C, Yu, C.L, Wang, G.X. | | Deposit date: | 2017-11-08 | | Release date: | 2018-11-07 | | Last modified: | 2019-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Water molecules in protein-ligand interfaces. Evaluation of software tools and SAR comparison.
J. Comput. Aided Mol. Des., 33, 2019
|
|
1NEJ
 
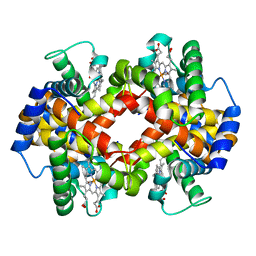 | | Crystalline Human Carbonmonoxy Hemoglobin S (Liganded Sickle Cell Hemoglobin) Exhibits The R2 Quaternary State At Neutral pH In The Presence Of Polyethylene Glycol: The 2.1 Angstrom Resolution Crystal Structure | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Patskovska, L.N, Patskovsky, Y.V, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2002-12-11 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | COHbC and COHbS crystallize in the R2 quaternary state at neutral pH in the presence of PEG 4000.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7ZZP
 
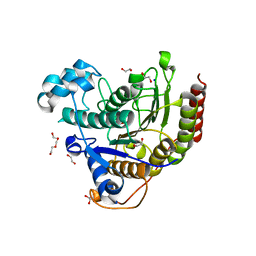 | | Structure of HDAC2 complexed with an inhibitory ligand | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
7ZZS
 
 | | HDAC2 complexed with an inhibitory ligand | | Descriptor: | (5~{S})-5-(4-chlorophenyl)pyrrolidin-2-one, 1,2-ETHANEDIOL, 2-(cyclohexylazaniumyl)ethanesulfonate, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
7ZZU
 
 | | Inhibitory Ligand binding to HDAC2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[(2~{R},4~{S})-4-phenylpyrrolidin-2-yl]carbonylpiperazin-1-yl]pyridine-3-carbonitrile, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cleasby, A, Tisi, D. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Based Discovery of a Novel, Brain Penetrant, Orally Active HDAC2 Inhibitor.
Acs Med.Chem.Lett., 13, 2022
|
|
4M4V
 
 | |
2A5T
 
 | | Crystal Structure Of The NR1/NR2A ligand-binding cores complex | | Descriptor: | GLUTAMIC ACID, GLYCINE, N-methyl-D-aspartate receptor NMDAR1-4a subunit, ... | | Authors: | Furukawa, H, Singh, S.K, Mancusso, R, Gouaux, E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-11-15 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subunit arrangement and function in NMDA receptors
Nature, 438, 2005
|
|
2XOZ
 
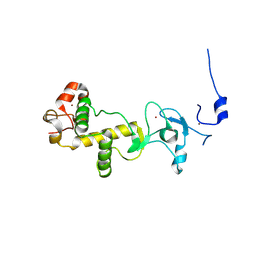 | | C-terminal cysteine rich domain of human CHFR bound to AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, E3 UBIQUITIN-PROTEIN LIGASE CHFR, ZINC ION | | Authors: | Oberoi, J, Bayliss, R. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Structural Basis of Poly(Adp-Ribose) Recognition by the Multizinc Binding Domain of Checkpoint with Forkhead-Associated and Ring Domains (Chfr).
J.Biol.Chem., 285, 2010
|
|
6BQA
 
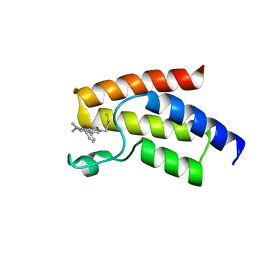 | | BRD9 bromodomain in complex with 3-(6-(but-3-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)-N,N-dimethylbenzamide | | Descriptor: | 3-[6-(but-3-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl]-N,N-dimethylbenzamide, Bromodomain-containing protein 9 | | Authors: | Murray, J.M, Tang, Y. | | Deposit date: | 2017-11-27 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.031 Å) | | Cite: | Water molecules in protein-ligand interfaces. Evaluation of software tools and SAR comparison.
J. Comput. Aided Mol. Des., 33, 2019
|
|
4WRE
 
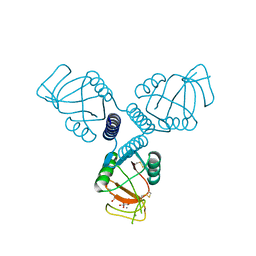 | | Crystal Structure of Surfactant Protein-A DEDN Mutant (E171D/P175E/R197N/K203D) Complexed with Inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Rynkiewicz, M.J, Wu, H, Cafarella, T.R, Nikolaidis, N.M, Head, J.F, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2014-10-23 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop
To be published
|
|
4WR9
 
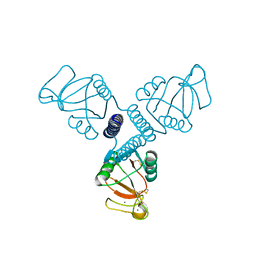 | | Crystal Structure of Surfactant Protein-A DED Mutant (E171D/P175E/K203D) | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A | | Authors: | Rynkiewicz, M.J, Wu, H, Cafarella, T.R, Nikolaidis, N.M, Head, J.F, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2014-10-23 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop
To be published
|
|
4WRC
 
 | | Crystal Structure of Surfactant Protein-A DEDN Mutant (E171D/P175E/R197N/K203D) | | Descriptor: | CALCIUM ION, CHLORIDE ION, HEXANE-1,6-DIOL, ... | | Authors: | Rynkiewicz, M.J, Wu, H, Cafarella, T.R, Nikolaidis, N.M, Head, J.F, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2014-10-23 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop
to be published
|
|
4WUX
 
 | | Crystal Structure of Surfactant Protein-A DED Mutant (E171D/P175E/K203D) Complexed with Mannose | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A, alpha-D-mannopyranose | | Authors: | Rynkiewicz, M.J, Wu, H, Cafarella, T.R, Nikolaidis, N.M, Head, J.F, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2014-11-03 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop
To be published
|
|
3RNN
 
 | |
6BLN
 
 | | BTK complex with compound 13 | | Descriptor: | GLYCEROL, N-(3-{5-[(1,5-dimethyl-1H-pyrazol-3-yl)amino]-6-oxo-1,6-dihydropyridazin-3-yl}-2,6-difluorophenyl)-4,5,6,7-tetrahydro-1-benzothiophene-2-carboxamide, SULFATE ION, ... | | Authors: | Kiefer, J.R, Eigenbrot, C, Yu, C.L. | | Deposit date: | 2017-11-10 | | Release date: | 2018-11-07 | | Last modified: | 2019-03-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Water molecules in protein-ligand interfaces. Evaluation of software tools and SAR comparison.
J. Comput. Aided Mol. Des., 33, 2019
|
|
6BQD
 
 | | TAF1-BD2 bromodomain in complex with (E)-3-(6-(but-2-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)-N,N-dimethylbenzamide | | Descriptor: | 3-{6-[(2E)-but-2-en-1-yl]-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl}-N,N-dimethylbenzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Murray, J.M, Tang, Y. | | Deposit date: | 2017-11-27 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.136 Å) | | Cite: | Water molecules in protein-ligand interfaces. Evaluation of software tools and SAR comparison.
J. Comput. Aided Mol. Des., 33, 2019
|
|
4WUW
 
 | | Crystal Structure of Surfactant Protein-A DED Mutant (E171D/P175E/K203D) Complexed with Inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, CALCIUM ION, Pulmonary surfactant-associated protein A | | Authors: | Rynkiewicz, M.J, Wu, H, Cafarella, T.R, Nikolaidis, N.M, Head, J.F, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2014-11-03 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Differential ligand binding specificities of the pulmonary collectins are determined by the conformational freedom of a surface loop
To be published
|
|
5X2M
 
 | | Crystal structure of the medaka fish taste receptor T1r2a-T1r3 ligand binding domains in complex with L-glutamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Nuemket, N, Yasui, N, Atsumi, N, Yamashita, A. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural basis for perception of diverse chemical substances by T1r taste receptors
Nat Commun, 8, 2017
|
|
